7US4
 
 | | Sars-Cov2 Main Protease in complex with CDD-1819 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M.M, Judge, A. | | Deposit date: | 2022-04-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
7UR9
 
 | | SARS-Cov2 Main protease in complex with inhibitor CDD-1845 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[4-(methylamino)-4-oxobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
7URB
 
 | | Sars-Cov2 Main Protease in complex with CDD-1733 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-{(1S)-1-[4-(trifluoromethyl)phenyl]butyl}-1H-benzimidazole-7-carboxamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M.M, Judge, A. | | Deposit date: | 2022-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
8CYR
 
 | | Alpha-synuclein fibril from spontaneous control | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Distinct cryo-EM structures and functions of alpha-synuclein fibrils amplified from cerebrospinal fluid
To Be Published
|
|
7XKE
 
 | | Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKF
 
 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKD
 
 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
6AEZ
 
 | | Crystal structure of human CCL5 trimer | | Descriptor: | C-C motif chemokine 5, SULFATE ION | | Authors: | Chen, Y.C, Li, K.M, Chen, P.J, Zarivach, R, Sun, Y.J, Sue, S.C. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Integrative Model to Coordinate the Oligomerization and Aggregation Mechanisms of CCL5.
J.Mol.Biol., 432, 2020
|
|
5YAM
 
 | |
5YC7
 
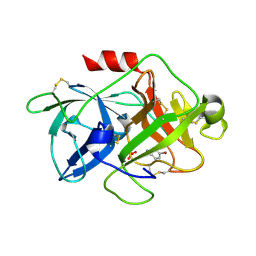 | |
5Z1C
 
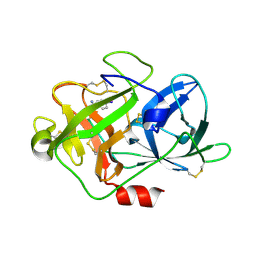 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | Descriptor: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | Deposit date: | 2017-12-25 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
6A8G
 
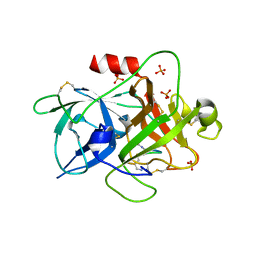 | | The crystal structure of muPAin-1-IG in complex with muPA-SPD at pH8.5 | | Descriptor: | PHOSPHATE ION, Urokinase-type plasminogen activator chain B, muPAin-1-IG | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-08 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
6A8N
 
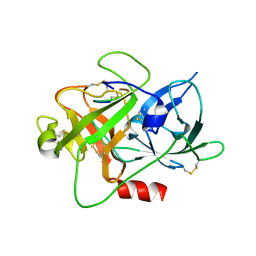 | | The crystal structure of muPAin-1-IG-2 in complex with muPA-SPD at pH8.5 | | Descriptor: | CYS-PRO-ALA-TYR-SER-ARG-TYR-ILE-GLY-CYS, Urokinase-type plasminogen activator B | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-09 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
2FDB
 
 | |
