8R5N
 
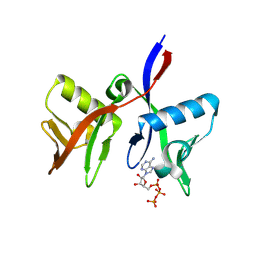 | | DTX1 WWE domain in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, E3 ubiquitin-protein ligase DTX1 | | Authors: | Muenzker, L, Zak, K.M, Boettcher, J. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8R6B
 
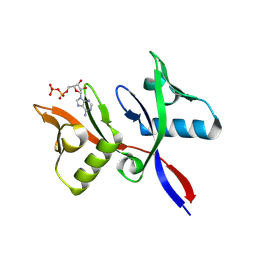 | |
8R7O
 
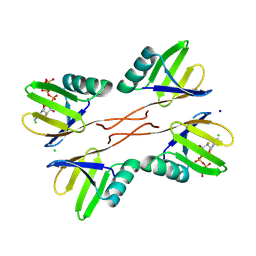 | | HUWE1 WWE domain in complex with 2'F-ATP | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1, SODIUM ION, ... | | Authors: | Muenzker, L, Boettcher, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8RD1
 
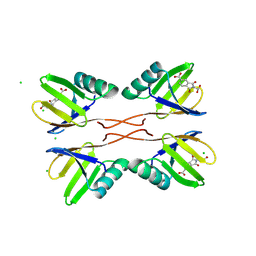 | | HUWE1 WWE domain in complex with compound 4 | | Descriptor: | 2-(2-hydroxy-2-oxoethyl)-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid, CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Muenzker, L, Zak, K.M, Boettcher, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
8R6A
 
 | |
8RD0
 
 | | HUWE1 WWE domain in complex with compound 3 | | Descriptor: | (1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetic acid, CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Muenzker, L, Boettcher, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
2C0O
 
 | | Src family kinase Hck with bound inhibitor A-770041 | | Descriptor: | CALCIUM ION, N-(4-{1-[4-(4-ACETYLPIPERAZIN-1-YL)-TRANS-CYCLOHEXYL]-4-AMINO-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL}-2-METHOXYPHENYL)-1-METHYL-1H-INDOLE-2-CARBOXAMIDE, TYROSINE-PROTEIN KINASE HCK | | Authors: | Borhani, D.W, Burchat, A, Calderwood, D.J, Hirst, G.C, Li, B, Loew, A. | | Deposit date: | 2005-09-06 | | Release date: | 2006-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of A-770041, a Src-Family Selective Orally Active Lck Inhibitor that Prevents Organ Allograft Rejection.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2C0I
 
 | | Src family kinase Hck with bound inhibitor A-420983 | | Descriptor: | CALCIUM ION, N-(4-{4-AMINO-1-[4-(4-METHYLPIPERAZIN-1-YL)-TRANS-CYCLOHEXYL]-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL}-2-METHOXYPHENYL)-1-METHYL-1H-INDOLE-2-CARBOXAMIDE, TYROSINE-PROTEIN KINASE HCK | | Authors: | Borhani, D.W, Burchat, A, Calderwood, D.J, Hirst, G.C, Li, B, Loew, A. | | Deposit date: | 2005-09-03 | | Release date: | 2006-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of A-770041, a Src-Family Selective Orally Active Lck Inhibitor that Prevents Organ Allograft Rejection.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2C0T
 
 | | Src family kinase Hck with bound inhibitor A-641359 | | Descriptor: | CALCIUM ION, N-(4-{4-AMINO-1-[1-(TETRAHYDRO-2H-PYRAN-4-YL)PIPERIDIN-4-YL]-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL}-2-METHOXYPHENYL)-1-METHYL-1H-INDOLE-2-CARBOXAMIDE, TYROSINE-PROTEIN KINASE HCK | | Authors: | Borhani, D.W, Burchat, A, Calderwood, D.J, Hirst, G.C, Li, B, Loew, A. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of A-770041, a Src-Family Selective Orally Active Lck Inhibitor that Prevents Organ Allograft Rejection.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7SZ2
 
 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
7SZ3
 
 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ADPr | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
6B09
 
 | |
5WJI
 
 | | Crystal structure of the F61S mutant of HsNUDT16 | | Descriptor: | ACETIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-07-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5W6Z
 
 | | Crystal structure of the H24W mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5VY2
 
 | | Crystal structure of the F36A mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-05-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5W6X
 
 | |
