5VQE
 
 | | Beta-glucoside phosphorylase BglX bound to 2FGlc | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucoside phosphorylase BglX | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
5VQD
 
 | | Beta-glucoside phosphorylase BglX | | Descriptor: | Beta-glucoside phosphorylase BglX, GLYCEROL | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
8A24
 
 | |
8A25
 
 | | Lysophospholipase PlaA from Legionella pneumophila str. Corby - complex with PEG fragment | | Descriptor: | (R,R)-2,3-BUTANEDIOL, IODIDE ION, Lysophospholipase A, ... | | Authors: | Diwo, M.G, Blankenfeldt, W. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-function relationships underpin disulfide loop cleavage-dependent activation of Legionella pneumophila lysophospholipase A PlaA.
Mol.Microbiol., 121, 2024
|
|
8A26
 
 | | Lysophospholipase PlaA from Legionella pneumophila str. Corby - complex with palmitate | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Lysophospholipase A, MALONIC ACID, ... | | Authors: | Diwo, M.G, Blankenfeldt, W. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-function relationships underpin disulfide loop cleavage-dependent activation of Legionella pneumophila lysophospholipase A PlaA.
Mol.Microbiol., 121, 2024
|
|
5FHX
 
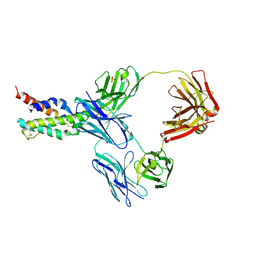 | | CRYSTAL STRUCTURE OF CODV IN COMPLEX WITH IL4 AT 2.55 Ang. RESOLUTION. | | Descriptor: | Antibody fragment light chain, Interleukin-4, PHOSPHATE ION, ... | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
5HCG
 
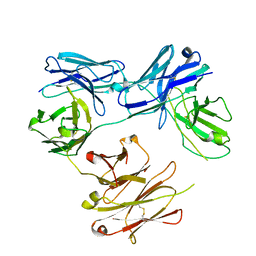 | | CRYSTAL STRUCTURE OF FREE-CODV. | | Descriptor: | CODV heavy-chain, CODV light chain | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
5WHZ
 
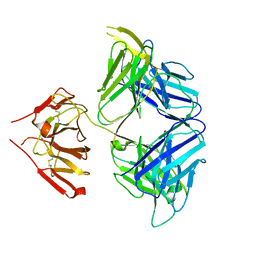 | | PGDM1400-10E8v4 CODV Fab | | Descriptor: | Anti-HIV CODV-Fab Heavy chain, Anti-HIV CODV-Fab Light chain | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.549 Å) | | Cite: | Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques.
Science, 358, 2017
|
|
1OX4
 
 | |
7B4S
 
 | | Structure of the 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus in complex with Coenzyme A and compound 153786 from the NCI Open Database | | Descriptor: | 5-[(4-chlorophenyl)methyl]-6-[[2-(dimethylamino)ethylamino]methyl]-4-oxidanyl-1~{H}-pyrimidine-2-thione, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Maveyraud, L, Carivenc, C, Mourey, L. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
7BDW
 
 | | Crystal structure of mycobacterial PptAb in complex with ACP and compound 8918 | | Descriptor: | COENZYME A, MANGANESE (II) ION, N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea, ... | | Authors: | Carivenc, C, Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2020-12-22 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
7BCZ
 
 | | Structure of the 4'-phosphopantetheinyl transferase PcpS from Pseudomonas aeruginosa | | Descriptor: | 4'-phosphopantetheinyl transferase EntD, COENZYME A, MAGNESIUM ION, ... | | Authors: | Carivenc, C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2020-12-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
4J93
 
 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BI-1 | | Descriptor: | (4S)-3-phenyl-4-(pyridin-3-yl)-4,5-dihydropyrrolo[3,4-c]pyrazol-6(2H)-one, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein | | Authors: | Lemke, C.T. | | Deposit date: | 2013-02-15 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Novel Small-Molecule HIV-1 Replication Inhibitors That Stabilize Capsid Complexes.
Antimicrob.Agents Chemother., 57, 2013
|
|
8CX0
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC monomeric complex | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX2
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 2 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX1
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8C03
 
 | | Structure of SLC40/ferroportin in complex with vamifeport and synthetic nanobody Sy12 in outward-facing conformation | | Descriptor: | 2-[2-[2-(1~{H}-benzimidazol-2-yl)ethylamino]ethyl]-~{N}-[(3-fluoranylpyridin-2-yl)methyl]-1,3-oxazole-4-carboxamide, Solute carrier family 40 member 1 | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
8BZY
 
 | | Structure of SLC40/ferroportin in complex with vamifeport and synthetic nanobody Sy3 in occluded conformation | | Descriptor: | 2-[2-[2-(1~{H}-benzimidazol-2-yl)ethylamino]ethyl]-~{N}-[(3-fluoranylpyridin-2-yl)methyl]-1,3-oxazole-4-carboxamide, DIUNDECYL PHOSPHATIDYL CHOLINE, Solute carrier family 40 member 1, ... | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
8C02
 
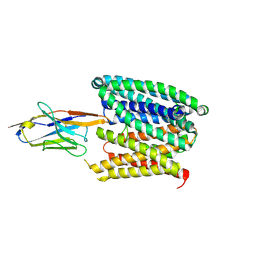 | | Structure of SLC40/ferroportin in complex with synthetic nanobody Sy3 in occluded conformation | | Descriptor: | Solute carrier family 40 member 1, Sybody3 | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
7K7Q
 
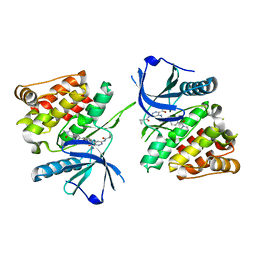 | |
7K7O
 
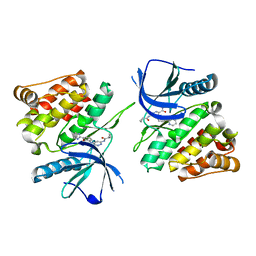 | |
3FZZ
 
 | | Structure of GrC | | Descriptor: | Granzyme C, SULFATE ION | | Authors: | Buckle, A.M, Kaiserman, D, Whisstock, J.C. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of granzyme C reveals an unusual mechanism of protease autoinhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2JTX
 
 | |
5VM7
 
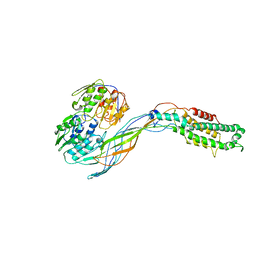 | | Pseudo-atomic model of the MurA-A2 complex | | Descriptor: | Maturation protein A2, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6OAT
 
 | | Structure of the Ganjam virus OTU bound to sheep ISG15 | | Descriptor: | Interferon stimulated gene 17, RNA-dependent RNA polymerase, prop-2-en-1-amine | | Authors: | Dzimianski, J.V, Williams, I.L, Pegan, S.D. | | Deposit date: | 2019-03-18 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Determining the molecular drivers of species-specific interferon-stimulated gene product 15 interactions with nairovirus ovarian tumor domain proteases.
Plos One, 14, 2019
|
|
