6RZU
 
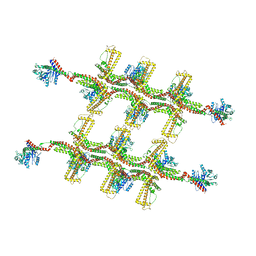 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZT
 
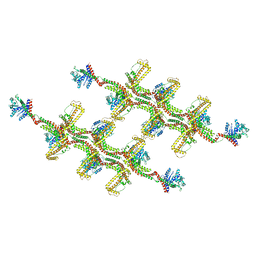 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
4HJ0
 
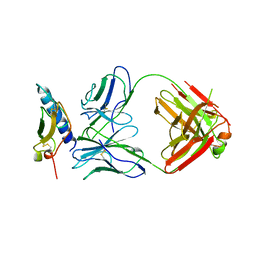 | | Crystal structure of the human GIPr ECD in complex with Gipg013 Fab at 3-A resolution | | Descriptor: | Gastric inhibitory polypeptide receptor, Gipg013 Fab, Antagonizing antibody to the GIP Receptor, ... | | Authors: | Madhurantakam, C, Ravn, P, Gruetter, M.G, Jackson, R.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Pharmacological Characterization of Novel Potent and Selective Monoclonal Antibody Antagonists of Glucose-dependent Insulinotropic Polypeptide Receptor.
J.Biol.Chem., 288, 2013
|
|
8OR4
 
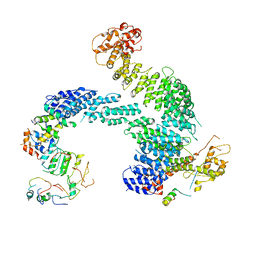 | | Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR3
 
 | | CAND1-CUL1-RBX1-SKP1-SKP2-DCNL1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR0
 
 | | CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR2
 
 | | CAND1-CUL1-RBX1-DCNL1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OUY
 
 | | Human RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complex, 3.4 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD51 homolog 2, ... | | Authors: | Greenhough, L.A, Liang, C.C, West, S.C. | | Deposit date: | 2023-04-25 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour suppressor.
Nature, 619, 2023
|
|
8OUZ
 
 | | Human RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complex, 2.2 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD51 homolog 2, ... | | Authors: | Greenhough, L.A, Liang, C.C, West, S.C. | | Deposit date: | 2023-04-25 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour suppressor.
Nature, 619, 2023
|
|
5AO1
 
 | |
5AO2
 
 | |
5AO0
 
 | |
5AO3
 
 | |
5AO4
 
 | |
1QUR
 
 | |
8QHQ
 
 | | Crystal structure of human DNPH1 bound to hmdUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
8QHR
 
 | | Crystal structure of the human DNPH1 glycosyl-enzyme intermediate | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, ... | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
8QXK
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State I - Tense | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXO
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State V - Depleted relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXN
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State IV - Depleted relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXL
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State II - Hemi-relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXM
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State III - Relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXJ
 
 | | Cryo-EM structure of tetrameric human SAMHD1 with dApNHpp | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
5C4K
 
 | | APH(2")-IVa in complex with GET (G418) at room temperature | | Descriptor: | APH(2'')-Id, GENETICIN | | Authors: | Kaplan, E, Guichou, J.F, Berrou, K, Chaloin, L, Leban, N, Lallemand, P, Barman, T, Serpersu, E.H, Lionne, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Aminoglycoside binding and catalysis specificity of aminoglycoside 2-phosphotransferase IVa: A thermodynamic, structural and kinetic study.
Biochim.Biophys.Acta, 1860, 2016
|
|
5C4L
 
 | | Conformational alternate of sisomicin in complex with APH(2")-IVa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, (2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-ol, APH(2'')-Id | | Authors: | Kaplan, E, Guichou, J.F, Berrou, K, Chaloin, L, Leban, N, Lallemand, P, Barman, T, Serpersu, E.H, Lionne, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aminoglycoside binding and catalysis specificity of aminoglycoside 2-phosphotransferase IVa: A thermodynamic, structural and kinetic study.
Biochim.Biophys.Acta, 1860, 2016
|
|
