3O5X
 
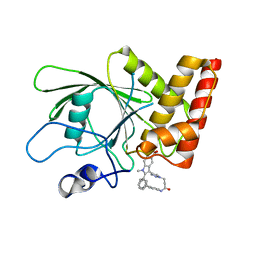 | | Crystal structure of the oncogenic tyrosine phosphatase SHP2 complexed with a salicylic acid-based small molecule inhibitor | | Descriptor: | 3-{1-[3-(biphenyl-4-ylamino)-3-oxopropyl]-1H-1,2,3-triazol-4-yl}-6-hydroxy-1-methyl-2-phenyl-1H-indole-5-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Zhang, X, He, Y, Liu, S, Yu, Z, Jiang, Z, Yang, Z, Dong, Y, Nabinger, S.C, Wu, L, Gunawan, A.M, Wang, L, Chan, R.J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2).
J.Med.Chem., 53, 2010
|
|
4LYR
 
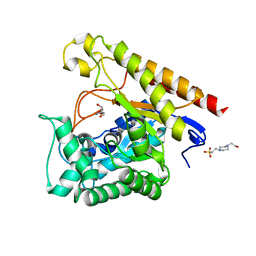 | | Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LYQ
 
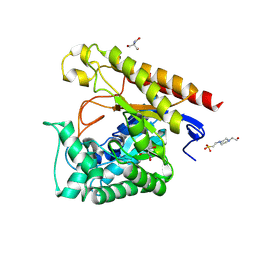 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase, ... | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3RN5
 
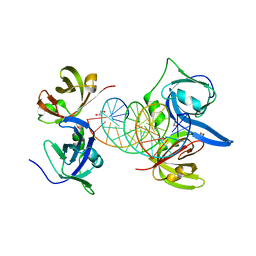 | | Structural basis of cytosolic DNA recognition by innate immune receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*AP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*TP*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*G)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RNU
 
 | | Structural Basis of Cytosolic DNA Sensing by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RN2
 
 | | Structural Basis of Cytosolic DNA Recognition by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*TP*CP*TP*TP*TP*GP*AP*TP*GP*G)-3'), Interferon-inducible protein AIM2 | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RLN
 
 | |
3RLO
 
 | | Structural Basis of Cytosolic DNA Recognition by Innate Receptors | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Gamma-interferon-inducible protein 16 | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
6KFW
 
 | |
7BUJ
 
 | | mcGAS bound with pppGpG | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
7BUM
 
 | | mcGAS bound with pGpA | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
7BUQ
 
 | | mcGAS bound with 23-cGAMP | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
4M58
 
 | | Crystal Structure of an transition metal transporter | | Descriptor: | Cobalamin biosynthesis protein CbiM, NICKEL (II) ION | | Authors: | Yu, Y, Yan, C.Y, Zhang, B, Li, X.L, Gu, J.K. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
4M5B
 
 | | Crystal Structure of an Truncated Transition Metal Transporter | | Descriptor: | Cobalamin biosynthesis protein CbiM, HEXANE-1,6-DIOL, NICKEL (II) ION | | Authors: | Yu, Y. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
4QP0
 
 | | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei | | Descriptor: | Endo-beta-mannanase, SULFATE ION | | Authors: | Zheng, Q.J, Peng, Z, Liu, Y, Yan, Q.J, Chen, Z.Z, Qin, Z. | | Deposit date: | 2014-06-22 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5C3U
 
 | | Crystal structure of a fungal L-serine ammonia-lyase from Rhizomucor miehei | | Descriptor: | L-serine ammonia-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhen, Q, Qiaojuan, Y, Shaoqing, Y, Zhengqiang, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure and characterization of a novel l-serine ammonia-lyase from Rhizomucor miehei.
Biochem.Biophys.Res.Commun., 466, 2015
|
|
7RMR
 
 | | Crystal structure of [I11L]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7RMQ
 
 | | Crystal structure of cycloviolacin O2 | | Descriptor: | Cycloviolacin O2, D-[I11L]cycloviolacin O2, FORMIC ACID | | Authors: | Huang, Y.H, Du, Q. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7RMS
 
 | | Crystal structure of [I11G]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
4LYP
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NRR
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-beta-D-fructofuranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NRS
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7SQI
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C14-crypto Acyl Carrier Protein, AcpP | | Descriptor: | Acyl carrier protein, Beta-ketoacyl-ACP synthase I, N-{2-[(2Z)-3-chlorotetradec-2-enamido]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Chen, A, Mindrebo, J.T, Davis, T.D, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-05 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SZ9
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16:1-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-(2-{[(9Z)-hexadec-9-enoyl]amino}ethyl)-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Chen, A, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5YLH
 
 | | Structure of GH113 beta-1,4-mannanase | | Descriptor: | beta-1,4-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
