7A02
 
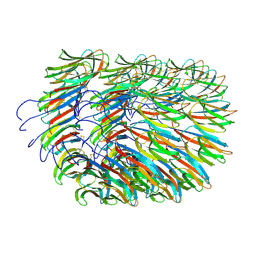 | | Bacillus endospore appendages form a novel family of disulfide-linked pili | | Descriptor: | DUF3992 domain-containing protein | | Authors: | Pradhan, B, Liedtke, J, Sleutel, M, Lindback, T, Llarena, A.K, Brynildsrud, O, Aspholm, M, Remaut, H. | | Deposit date: | 2020-08-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Endospore Appendages: a novel pilus superfamily from the endospores of pathogenic Bacilli.
Embo J., 40, 2021
|
|
7RK6
 
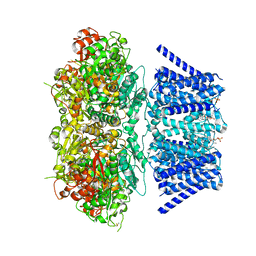 | | Aplysia Slo1 with Barium | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, BARIUM ION, BK channel, ... | | Authors: | Zhu, J, Srivastava, S, Cachau, R, Holmgren, M. | | Deposit date: | 2021-07-22 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | CryoEM structure of Aplysia Slo1 with 0 mM Ba2+ at 2.91 A
To Be Published
|
|
7RJT
 
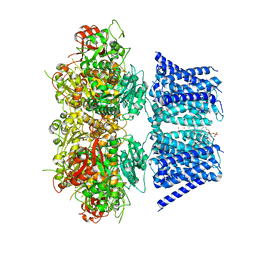 | | Aplysia Slo1 with Barium | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, BARIUM ION, BK channel, ... | | Authors: | Zhu, J, Srivastava, S, Cachau, R, Holmgren, M. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | CryoEM structure of Aplysia Slo1 with 10 mM Ba2+ at 2.93 A
To Be Published
|
|
7OTC
 
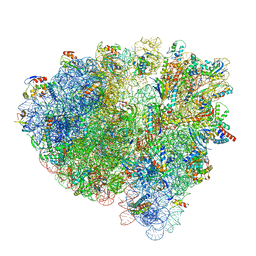 | | Cryo-EM structure of an Escherichia coli 70S ribosome in complex with elongation factor G and the antibiotic Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Wieland, M, Koller, T.O, Wilson, D.N. | | Deposit date: | 2021-06-10 | | Release date: | 2022-05-11 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6HHQ
 
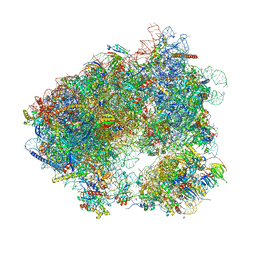 | | Crystal structure of compound C45 bound to the yeast 80S ribosome | | Descriptor: | (3~{R})-3-[(1~{S})-2-[(1~{S},4~{a}~{R},6~{S},7~{S},8~{a}~{R})-6,7-bis(chloranyl)-5,5,8~{a}-trimethyl-2-methylidene-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Pellegrino, S, Vanderwal, C.D, Yusupov, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.10000038 Å) | | Cite: | Understanding the role of intermolecular interactions between lissoclimides and the eukaryotic ribosome.
Nucleic Acids Res., 47, 2019
|
|
8P4V
 
 | | 80S yeast ribosome in complex with HaterumaimideQ | | Descriptor: | (3~{R})-3-[(1~{S})-2-[(1~{R},3~{S},4~{a}~{S},8~{a}~{S})-5,5,8~{a}-trimethyl-2-methylidene-3-oxidanyl-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Terrosu, S, Yusupov, M. | | Deposit date: | 2023-05-23 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Synthesis of Differentially Halogenated Lissoclimide Analogues To Probe Ribosome E-Site Binding.
Acs Chem.Biol., 20, 2025
|
|
8P9A
 
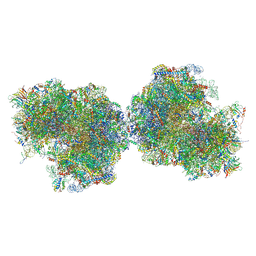 | | 80S yeast ribosome in complex with Methyllissoclimide | | Descriptor: | (3~{R})-3-[(1~{S})-2-[(1~{R},3~{S},4~{a}~{S},7~{S},8~{a}~{S})-5,5,7,8~{a}-tetramethyl-2-methylidene-3-oxidanyl-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 16S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Terrosu, S, Yusupov, M, Vanderwal, C. | | Deposit date: | 2023-06-05 | | Release date: | 2024-11-27 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis of Differentially Halogenated Lissoclimide Analogues To Probe Ribosome E-Site Binding.
Acs Chem.Biol., 20, 2025
|
|
8PNN
 
 | | 80S yeast ribosome in complex with Bromolissoclimide | | Descriptor: | (3~{R})-3-[(1~{S})-2-[(1~{R},3~{S},4~{a}~{S},7~{S},8~{a}~{S})-7-bromanyl-5,5,8~{a}-trimethyl-2-methylidene-3-oxidanyl-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 16S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Terrosu, S, Yusupov, M. | | Deposit date: | 2023-06-30 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis of Differentially Halogenated Lissoclimide Analogues To Probe Ribosome E-Site Binding.
Acs Chem.Biol., 20, 2025
|
|
8T9X
 
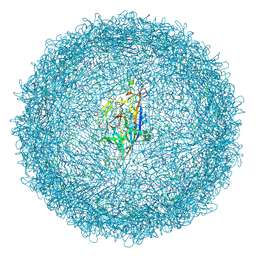 | |
8TA7
 
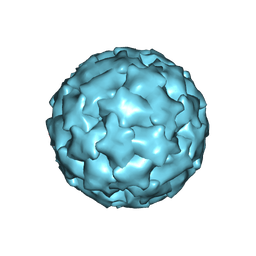 | |
8T9E
 
 | |
8T9C
 
 | |
8TJE
 
 | |
8BTM
 
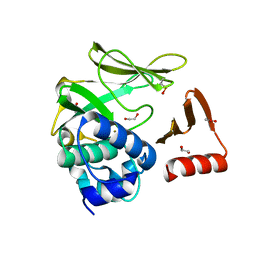 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GdmF | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-12 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of the geldanamycin amide synthase from Streptomyces hygroscopicus.
Nat Commun, 16, 2025
|
|
8OOM
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-{3-oxo-3-[(2-sulfanylethyl)amino]propyl}butanamide, ACETATE ION, GdmF | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-17 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure and function of the geldanamycin amide synthase from Streptomyces hygroscopicus.
Nat Commun, 16, 2025
|
|
8OSZ
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 3-aminophenol, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-05-01 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure and function of the geldanamycin amide synthase from Streptomyces hygroscopicus.
Nat Commun, 16, 2025
|
|
8OSV
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-05-01 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure and function of the geldanamycin amide synthase from Streptomyces hygroscopicus.
Nat Commun, 16, 2025
|
|
2A0M
 
 | |
3M3I
 
 | |
2Q0X
 
 | |
8VXE
 
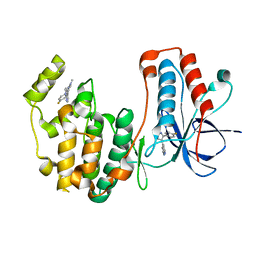 | | Structure of p38 alpha (Mitogen-activated protein kinase 14) complexed with inhibitor 6 | | Descriptor: | (4M)-4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Blaesse, M, Steinbacher, S, Shaffer, P.L, Sharma, S, Thompson, A.A. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
8VXD
 
 | |
8VXF
 
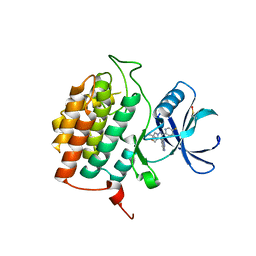 | | Structure of Casein kinase I isoform delta (CK1d) complexed with inhibitor 15 | | Descriptor: | (2P,3P,8S)-2-(5-fluoropyridin-2-yl)-6,6-dimethyl-3-(1H-pyrazolo[3,4-b]pyridin-4-yl)-6,7-dihydro-4H-pyrazolo[5,1-c][1,4]oxazine, Casein kinase I isoform delta | | Authors: | Thompson, A.A, Milligan, C.M, Sharma, S. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
8PDZ
 
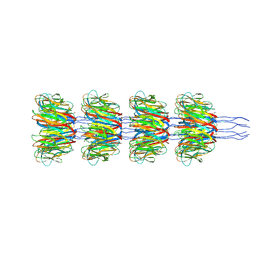 | | Recombinant Ena3A L-Type endospore appendages | | Descriptor: | DUF3992 domain-containing protein | | Authors: | Sleutel, M, Remaut, H. | | Deposit date: | 2023-06-13 | | Release date: | 2023-11-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A novel class of ultra-stable endospore appendages decorated with collagen-like tip fibrillae
Biorxiv, 2023
|
|
9HZE
 
 | |
