8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | Descriptor: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
2AJD
 
 | | Porcine dipeptidyl peptidase IV (CD26) in complex with L-Pro-boro-L-Pro (boroPro) | | Descriptor: | (2R)-N-[(2R)-2-(DIHYDROXYBORYL)-1-L-PROLYLPYRROLIDIN-2-YL]-N-[(5R)-5-(DIHYDROXYBORYL)-1-L-PROLYLPYRROLIDIN-2-YL]-L-PROLINAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
2AJC
 
 | | Porcine dipeptidyl peptidase IV (CD26) in complex with 4-(2-Aminoethyl)-benzene sulphonyl fluoride (AEBSF) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
2AJ8
 
 | | Porcine dipeptidyl peptidase IV (CD26) in complex with 7-Benzyl-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-purine-2,6-dione (BDPX) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
7P1W
 
 | |
5NXY
 
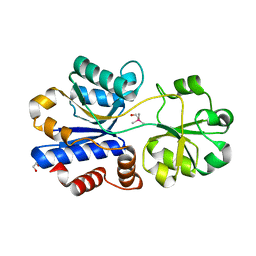 | | Crystal structure of OpuAC from B. subtilis in complex with Arsenobetaine | | Descriptor: | 1,2-ETHANEDIOL, 2-(trimethyl-lambda~5~-arsanyl)ethanol, Osmotically activated L-carnitine/choline ABC transporter substrate-binding protein OpuCC | | Authors: | Hofmann, T, Bremer, E, Schmitt, L, Smits, S. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes.
Environ. Microbiol., 20, 2018
|
|
5NXX
 
 | | Crystal structure of OpuAC from B. subtilis in complex with Arsenobetaine | | Descriptor: | (trimethylarsonio)acetate, Glycine betaine ABC transport system glycine betaine-binding protein OpuAC | | Authors: | Hofmann, T, Bremer, E, Schmitt, L, Smits, S. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes.
Environ. Microbiol., 20, 2018
|
|
7BM2
 
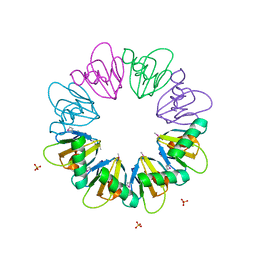 | |
7BME
 
 | |
6VSW
 
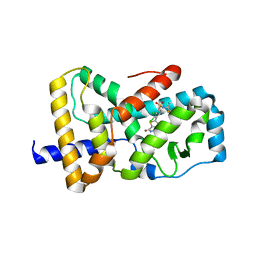 | | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of RORgt | | Descriptor: | 5-(2,3-dichloro-4-{[(2S)-1,1,1-trifluoropropan-2-yl]sulfamoyl}phenyl)-4-(4-fluoropiperidine-1-carbonyl)-N-(2-hydroxy-2-methylpropyl)-1,3-thiazole-2-carboxamide, RAR-related orphan receptor C | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2020-02-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of ROR gamma t.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7NYF
 
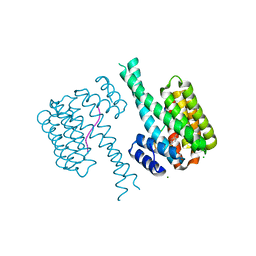 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-131 | | Descriptor: | 14-3-3 protein sigma, 4-[(3~{S})-3-oxidanylpiperidin-1-yl]sulfonylbenzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NZ6
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-125 | | Descriptor: | 14-3-3 protein sigma, 4-[4-(2-methoxyethyl)piperazin-1-yl]sulfonylbenzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-23 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NVI
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-186 | | Descriptor: | 14-3-3 protein sigma, 4-[(7-chloranyl-2,3-dihydro-1,4-benzoxazin-4-yl)sulfonyl]benzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-15 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O3F
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-117 | | Descriptor: | 1-methyl-4-(4-methylphenyl)sulfonyl-1,4-diazepane, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NY4
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-130 | | Descriptor: | 14-3-3 protein sigma, 4-(3-oxidanylidenepiperazin-1-yl)sulfonylbenzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-20 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NZG
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-122 | | Descriptor: | 14-3-3 protein sigma, 4-(4-methoxypiperidin-1-yl)sulfonylbenzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O3P
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-116 | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-6-methoxy-1-(4-methylphenyl)sulfonyl-benzimidazole, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O57
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-157 | | Descriptor: | 14-3-3 protein sigma, 3-morpholin-4-yl-4-nitro-benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O5D
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-160 | | Descriptor: | 14-3-3 protein sigma, 4-methanoyl-~{N}-[(4-methoxyphenyl)methyl]benzamide, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O5U
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-168 | | Descriptor: | 14-3-3 protein sigma, 4-methanoyl-~{N}-[(1-methylimidazol-2-yl)methyl]benzamide, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-09 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O6K
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-080 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) 3-chloranylbenzoate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NXY
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-181 | | Descriptor: | 14-3-3 protein sigma, 4-[(6-fluoranyl-3,4-dihydro-2~{H}-quinolin-1-yl)sulfonyl]benzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O3S
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-045 | | Descriptor: | 14-3-3 protein sigma, 3-methoxy-1-(4-methylphenyl)sulfonyl-azetidine, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O5S
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-167 | | Descriptor: | 14-3-3 protein sigma, 4-methanoyl-~{N}-[(3-methoxyphenyl)methyl]benzamide, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-09 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
