133D
 
 | | THE CRYSTAL STRUCTURE OF N4-METHYLCYTOSINE.GUANOSIN BASE-PAIRS IN THE SYNTHETIC HEXANUCLEOTIDE D(CGCGM(4)CG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C34)P*G)-3') | | Authors: | Cervi, A.R, Guy, A, Leonard, G.A, Teoule, R, Hunter, W.N. | | Deposit date: | 1993-07-29 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of N4-methylcytosine.guanosine base-pairs in the synthetic hexanucleotide d(CGCGm4CG).
Nucleic Acids Res., 21, 1993
|
|
1D75
 
 | | CONFORMATION OF THE GUANINE.8-OXOADENINE BASE PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(O8A)GCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(A38)P*GP*CP*G)-3') | | Authors: | Leonard, G.A, Guy, A, Brown, T, Teoule, R, Hunter, W.N. | | Deposit date: | 1992-05-07 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation of guanine-8-oxoadenine base pairs in the crystal structure of d(CGCGAATT(O8A)GCG).
Biochemistry, 31, 1992
|
|
9CKV
 
 | | Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Werther, R, Nguyen, A, Estrada Alamo, K.A, Wang, X, Campbell, M.G. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | De Novo Design of Integrin alpha5beta1 Modulating Proteins for Regenerative Medicine
To Be Published
|
|
1BEI
 
 | | Shk-dnp22: A Potent Kv1.3-specific immunosuppressive polypeptide, NMR, 20 structures | | Descriptor: | POTASSIUM CHANNEL TOXIN SHK | | Authors: | Kalman, K, Pennington, M.W, Lanigan, M.D, Nguyen, A, Rauer, H, Mahnir, V, Gutman, G.A, Paschetto, K, Kem, W.R, Grissmer, S, Christian, E.P, Cahalan, M.D, Norton, R.S, Chandy, K.G. | | Deposit date: | 1998-05-14 | | Release date: | 1998-12-02 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | ShK-Dap22, a potent Kv1.3-specific immunosuppressive polypeptide.
J.Biol.Chem., 273, 1998
|
|
2C1J
 
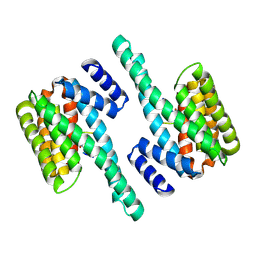 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-15 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
2C1N
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
7APL
 
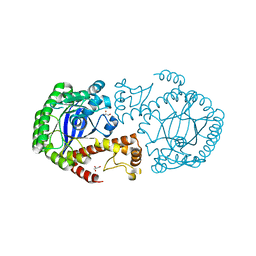 | |
7APM
 
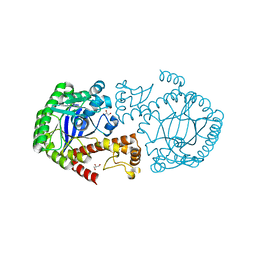 | | tRNA-guanine transglycosylase H319C mutant spin-labeled with MTSL. | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-10-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unraveling a Ligand-Induced Twist of a Homodimeric Enzyme by Pulsed Electron-Electron Double Resonance.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4RP7
 
 | |
4RP6
 
 | |
7JH5
 
 | | Co-LOCKR: de novo designed protein switch | | Descriptor: | Co-LOCKR: de novo designed protein switch | | Authors: | Bick, M.J, Lajoie, M.J, Boyken, S.E, Sankaran, B, Baker, D. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Designed protein logic to target cells with precise combinations of surface antigens.
Science, 369, 2020
|
|
6X00
 
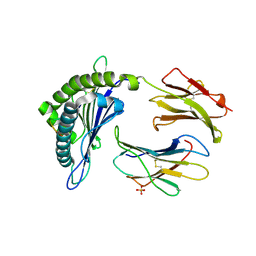 | | Structure of DbNA(11) peptides bound to H-2Db MHC-I | | Descriptor: | Beta-2-microglobulin, Epitope from Neuraminidase Protein (NA-181-191), H-2 class I histocompatibility antigen, ... | | Authors: | Farenc, C, Rossjohn, J. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Overlapping Peptides Elicit Distinct CD8 + T Cell Responses following Influenza A Virus Infection.
J Immunol., 205, 2020
|
|
6WZY
 
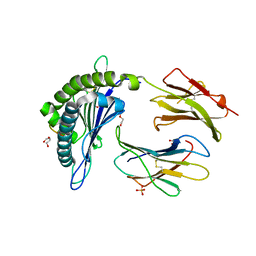 | | Structure of DbNA(10) peptides bound to H-2Db MHC-I | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Farenc, C, Rossjohn, J. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Overlapping Peptides Elicit Distinct CD8 + T Cell Responses following Influenza A Virus Infection.
J Immunol., 205, 2020
|
|
8AYH
 
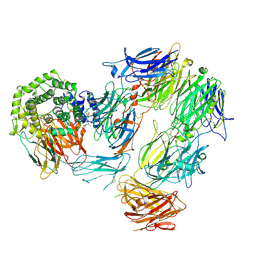 | |
8AXW
 
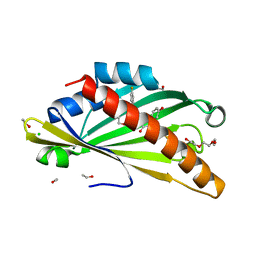 | | The structure of mouse AsterC (GramD1c) with Ezetimibe | | Descriptor: | (3~{R},4~{S})-1-(4-fluorophenyl)-3-[(3~{S})-3-(4-fluorophenyl)-3-oxidanyl-propyl]-4-(4-hydroxyphenyl)azetidin-2-one, CHLORIDE ION, ETHANOL, ... | | Authors: | Fairall, L, Xiao, X, Burger, L, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aster-dependent nonvesicular transport facilitates dietary cholesterol uptake.
Science, 382, 2023
|
|
7UV9
 
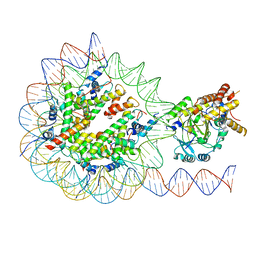 | | KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate | | Descriptor: | DNA (185-MER), FE (III) ION, Histone H2A type 1, ... | | Authors: | Spangler, C.J, Skrajna, A, Foley, C.A, Budziszewski, G.R, Azzam, D.N, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
7UVA
 
 | | Crystal structure of KDM2A histone demethylase catalytic domain in complex with an H3C36 peptide modified by UNC8015 | | Descriptor: | FE (III) ION, Histone H3.2, Lysine-specific demethylase 2A, ... | | Authors: | Budziszewski, G.R, Azzam, D.N, Spangler, C.J, Skrajna, A, Foley, C.A, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
3KHQ
 
 | | Crystal structure of murine Ig-beta (CD79b) in the monomeric form | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain, CITRATE ANION, GLUTATHIONE, ... | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-30 | | Release date: | 2010-08-25 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
3KG5
 
 | | Crystal structure of human Ig-beta homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-28 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
3KHO
 
 | | Crystal structure of murine Ig-beta (CD79b) homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain, SULFATE ION | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-30 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
3HR5
 
 | |
7JWI
 
 | |
7JWJ
 
 | | Crystal Structure of B17-C1 TCR-H2Db | | Descriptor: | B17.C1 TCR alpha chain, B17.C1 TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Farenc, C, Rossjohn, J, Gras, S, Szeto, C. | | Deposit date: | 2020-08-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Canonical T cell receptor docking on peptide-MHC is essential for T cell signaling.
Science, 372, 2021
|
|
3UIE
 
 | |
