2BWX
 
 | | His354Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWV
 
 | | His361Ala Escherichia coli Aminopeptidase P | | Descriptor: | AMINOPEPTIDASE P, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWS
 
 | | His243Ala Escherichia coli Aminopeptidase P | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, XAA-PRO AMINOPEPTIDASE P | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWT
 
 | | Asp260Ala Escherichia coli Aminopeptidase P | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRATE ANION, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BWW
 
 | | His350Ala Escherichia coli Aminopeptidase P | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, AMINOPEPTIDASE P, CITRATE ANION, ... | | Authors: | Graham, S.C, Guss, J.M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Kinetic and Crystallographic Analysis of Mutant Escherichia Coli Aminopeptidase P: Insights Into Substrate Recognition and the Mechanism of Catalysis.
Biochemistry, 45, 2006
|
|
2BH3
 
 | | Zn substituted E. coli Aminopeptidase P in complex with product | | Descriptor: | CITRATE ANION, LEUCINE, MAGNESIUM ION, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2005-01-07 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
4MJ3
 
 | | Haloalkane dehalogenase DmrA from Mycobacterium rhodesiae JS60 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, POTASSIUM ION | | Authors: | Fung, H, Gadd, M.S, Guss, J.M, Matthews, J.M. | | Deposit date: | 2013-09-03 | | Release date: | 2015-02-25 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and biophysical characterisation of haloalkane dehalogenases DmrA and DmrB in Mycobacterium strain JS60 and their role in growth on haloalkanes.
Mol.Microbiol., 97, 2015
|
|
2PCY
 
 | |
3PCY
 
 | | THE CRYSTAL STRUCTURE OF MERCURY-SUBSTITUTED POPLAR PLASTOCYANIN AT 1.9-ANGSTROMS RESOLUTION | | Descriptor: | MERCURY (II) ION, PLASTOCYANIN | | Authors: | Church, W.B, Guss, J.M, Potter, J.J, Freeman, H.C. | | Deposit date: | 1985-12-10 | | Release date: | 1986-01-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of mercury-substituted poplar plastocyanin at 1.9-A resolution.
J.Biol.Chem., 261, 1986
|
|
1DL8
 
 | | CRYSTAL STRUCTURE OF 5-F-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 5-FLUORO-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acridinecarboxamide topoisomerase poisons: structural and kinetic studies of the DNA complexes of 5-substituted 9-amino-(N-(2-dimethylamino)ethyl)acridine-4-carboxamides.
Mol.Pharmacol., 58, 2000
|
|
6CME
 
 | | Structure of wild-type ISL2-LID in complex with LHX4-LIM1+2 | | Descriptor: | LIM/homeobox protein Lhx4,Insulin gene enhancer protein ISL-2, ZINC ION | | Authors: | Stokes, P.H, Silva, A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins, 87, 2019
|
|
1PND
 
 | |
1PNC
 
 | |
4XS0
 
 | | Human methemoglobin in complex with the second and third NEAT domains of IsdH(F365Y/A369F/Y642A) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Dickson, C.F, Jacques, D.A, Guss, J.M, Gell, D.A. | | Deposit date: | 2015-01-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of haemoglobin bound to the haemoglobin receptor IsdH from Staphylococcus aureus shows disruption of the native alpha-globin haem pocket.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4F2J
 
 | | Crystal structure of ZNF217 bound to DNA, P6522 crystal form | | Descriptor: | 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', ZINC ION, Zinc finger protein 217 | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2012-05-08 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217
J.Biol.Chem., 288, 2013
|
|
5K6P
 
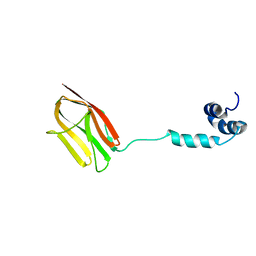 | | The NMR structure of the m domain tri-helix bundle and C2 of human cardiac Myosin Binding Protein C | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Michie, K.A, Kwan, A.H, Tung, C.S, Guss, J.M, Trewhella, J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure, 24, 2016
|
|
2V3Y
 
 | |
1M35
 
 | | Aminopeptidase P from Escherichia coli | | Descriptor: | AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Graham, S.C, Lee, M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-06-27 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthorhombic form of Escherichia coli aminopeptidase P at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2RGT
 
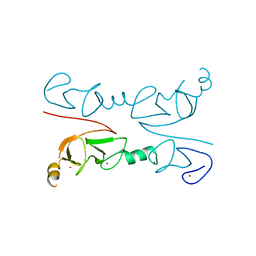 | | Crystal Structure of Lhx3 LIM domains 1 and 2 with the binding domain of Isl1 | | Descriptor: | Fusion of LIM/homeobox protein Lhx3, linker, Insulin gene enhancer protein ISL-1, ... | | Authors: | Bhati, M, Lee, M, Guss, J.M, Matthews, J.M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes.
Embo J., 27, 2008
|
|
4JCJ
 
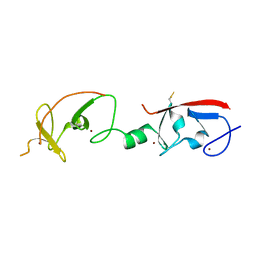 | | Crystal structure of Isl1 LIM domains with Ldb1 LIM-interaction domain | | Descriptor: | Insulin gene enhancer protein ISL-1,LIM domain-binding protein 1, ZINC ION | | Authors: | Gadd, M.S, Jacques, D.A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2013-02-21 | | Release date: | 2013-06-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural basis for the regulation of the LIM-homeodomain protein islet 1 (Isl1) by intra- and intermolecular interactions.
J.Biol.Chem., 288, 2013
|
|
1FSL
 
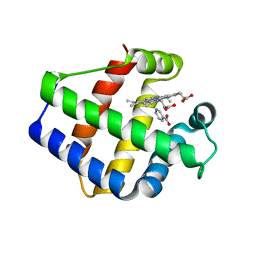 | | FERRIC SOYBEAN LEGHEMOGLOBIN COMPLEXED WITH NICOTINATE | | Descriptor: | LEGHEMOGLOBIN A, NICOTINIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ellis, P.J, Guss, J.M, Freeman, H.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ferric soybean leghemoglobin a nicotinate at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
4IS1
 
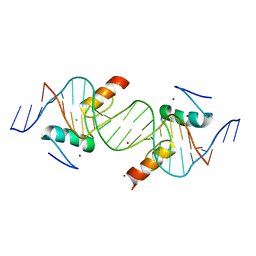 | | Crystal structure of ZNF217 bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', CHLORIDE ION, ... | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217.
J.Biol.Chem., 288, 2013
|
|
6OM5
 
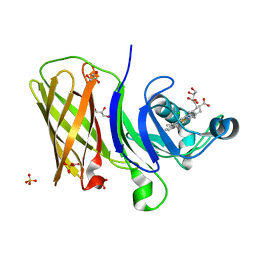 | | Structure of a haemophore from Haemophilus haemolyticus | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Torrado, M, Walshe, J.L, Mackay, J.P, Guss, J.M, Gell, D.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A heme-binding protein produced by Haemophilus haemolyticus inhibits non-typeable Haemophilus influenzae.
Mol.Microbiol., 113, 2020
|
|
3G9Y
 
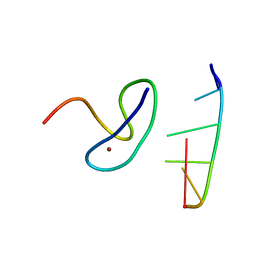 | | Crystal structure of the second zinc finger from ZRANB2/ZNF265 bound to 6 nt ssRNA sequence AGGUAA | | Descriptor: | RNA (5'-R(*AP*GP*GP*UP*AP*A)-3'), ZINC ION, Zinc finger Ran-binding domain-containing protein 2 | | Authors: | Loughlin, F.E, McGrath, A.P, Lee, M, Guss, J.M, Mackay, J.P. | | Deposit date: | 2009-02-15 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4FC3
 
 | | Crystal Structure of Human Methaemoglobin Complexed with the Second NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein H, ... | | Authors: | Krishna Kumar, K, Jacques, D.A, Guss, J.M, Gell, D.A. | | Deposit date: | 2012-05-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of the Hemoglobin-IsdH Complex Reveals the Molecular Basis of Iron Capture by Staphylococcus aureus
J.Biol.Chem., 289, 2014
|
|
