8WTJ
 
 | | XBB.1.5.70 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTD
 
 | | XBB.1.5.10 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8JPP
 
 | | Cryo-EM structure of succinate receptor bound to succinate acid coupling MiniGsq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-06-12 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 2024
|
|
8J22
 
 | | Cryo-EM structure of FFAR2 complex bound with TUG-1375 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J24
 
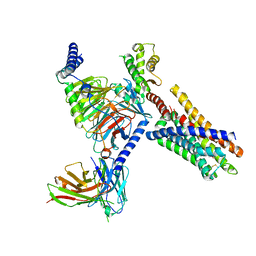 | | Cryo-EM structure of FFAR2 complex bound with acetic acid | | Descriptor: | ACETATE ION, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Tang, W, Sun, X, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J20
 
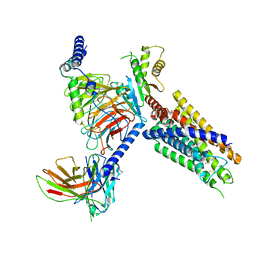 | | Cryo-EM structure of FFAR3 bound with valeric acid and AR420626 | | Descriptor: | (4R)-N-[2,5-bis(chloranyl)phenyl]-4-(furan-2-yl)-2-methyl-5-oxidanylidene-4,6,7,8-tetrahydro-1H-quinoline-3-carboxamide, Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J21
 
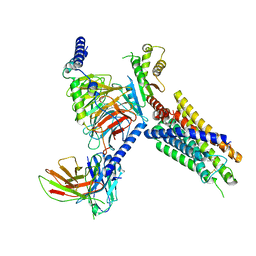 | | Cryo-EM structure of FFAR3 complex bound with butyrate acid | | Descriptor: | Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8JPN
 
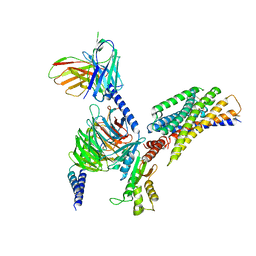 | | Cryo-EM structure of succinate receptor bound to cis-epoxysuccinic acid coupling to Gi | | Descriptor: | (2R,3S)-oxirane-2,3-dicarboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-06-12 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 2024
|
|
8K8J
 
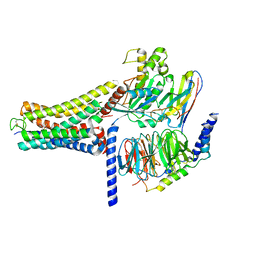 | | Cannabinoid Receptor 1 bound to Fenofibrate coupling MiniGsq and Nb35 Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Tang, W.Q, Wang, T.X, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Fenofibrate Recognition and G q Protein Coupling Mechanisms of the Human Cannabinoid Receptor CB1.
Adv Sci, 11, 2024
|
|
5ZNJ
 
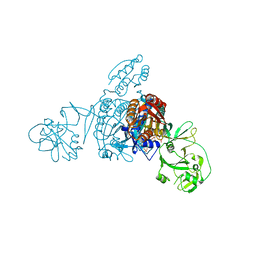 | | Crystal structure of a bacterial ProRS with ligands | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cheng, B, Yu, Y, Zhou, H. | | Deposit date: | 2018-04-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
5ZNK
 
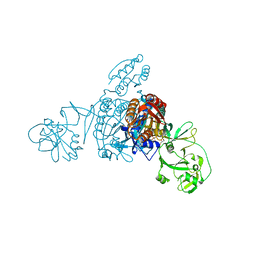 | | Crystal structure of a bacterial ProRS with ligands | | Descriptor: | 7-chloro-6-fluoro-3-{2-oxo-3-[(2S)-piperidin-2-yl]propyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cheng, B, Yu, Y, Zhou, H. | | Deposit date: | 2018-04-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
7XNS
 
 | | SARS-CoV-2 Omicron BA.2.12.1 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIY
 
 | | SARS-CoV-2 Omicron BA.3 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIX
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNQ
 
 | | SARS-CoV-2 Omicron BA.4 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIW
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIZ
 
 | | SARS-CoV-2 Omicron BA.3 variant spike (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNR
 
 | | SARS-CoV-2 Omicron BA.2.13 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7X6A
 
 | |
7YQT
 
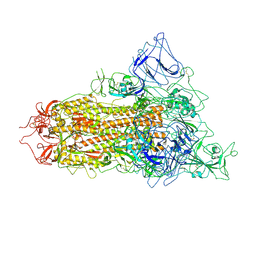 | | SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR3
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQV
 
 | | pH 5.5 SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR1
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with XG2v024 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR2
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQU
 
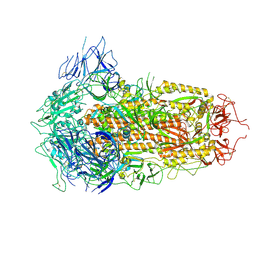 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
