2WVU
 
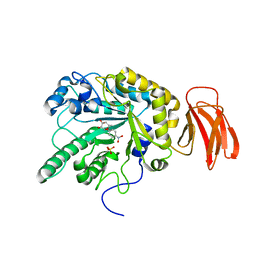 | | Crystal structure of a Michaelis complex of alpha-L-fucosidase GH29 from Bacteroides thetaiotaomicron with the synthetic substrate 4- nitrophenyl-alpha-L-fucose | | Descriptor: | 4-nitrophenyl 6-deoxy-alpha-L-galactopyranoside, ALPHA-L-FUCOSIDASE, SULFATE ION | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach
J.Am.Chem.Soc., 132, 2010
|
|
2WVT
 
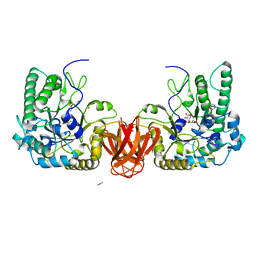 | | Crystal structure of an alpha-L-fucosidase GH29 from Bacteroides thetaiotaomicron in complex with a novel iminosugar fucosidase inhibitor | | Descriptor: | (2S,3R,5R,6S)-3,4,5-TRIHYDROXY-2,6-BIS(HYDROXYMETHYL)PIPERIDINIUM, ALPHA-L-FUCOSIDASE, GLYCEROL, ... | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach.
J.Am.Chem.Soc., 132, 2010
|
|
2WVV
 
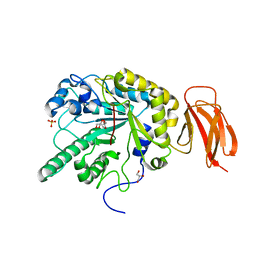 | | Crystal structure of an alpha-L-fucosidase GH29 from Bacteroides thetaiotaomicron | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-L-FUCOSIDASE, GLYCEROL, ... | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach
J.Am.Chem.Soc., 132, 2010
|
|
5FD5
 
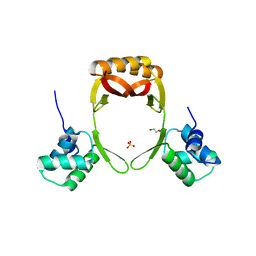 | | manganese uptake regulator | | Descriptor: | 1,2-ETHANEDIOL, Ferric uptake regulation protein, SULFATE ION | | Authors: | Bellini, D, Lebedev, A, Keegan, R, Walsh, M.A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of an apo metal-free manganese uptake regulator, mur
To Be Published
|
|
4CNU
 
 | |
1ZUA
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Tolrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT | | Authors: | Gallego, O, Ruiz, F.X, Ardevol, A, Dominguez, M, Alvarez, R, de Lera, A.R, Rovira, C, Farres, J, Fita, I, Pares, X. | | Deposit date: | 2005-05-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5HT6
 
 | | Crystal structure of the SET domain of the human MLL5 methyltransferase | | Descriptor: | Histone-lysine N-methyltransferase 2E | | Authors: | le Maire, A, Mas-y-Mas, S, Dumas, C, Lebedev, A. | | Deposit date: | 2016-01-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | The Human Mixed Lineage Leukemia 5 (MLL5), a Sequentially and Structurally Divergent SET Domain-Containing Protein with No Intrinsic Catalytic Activity.
Plos One, 11, 2016
|
|
2J5I
 
 | | Crystal Structure of Hydroxycinnamoyl-CoA Hydratase-Lyase | | Descriptor: | P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Leonard, P.M, Brzozowski, A.M, Lebedev, A, Marshall, C.M, Smith, D.J, Verma, C.S, Walton, N.J, Grogan, G. | | Deposit date: | 2006-09-18 | | Release date: | 2006-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A Resolution Structure of Hydroxycinnamoyl- Coenzyme a Hydratase-Lyase (Hchl) from Pseudomonas Fluorescens, an Enzyme that Catalyses the Transformation of Feruloyl-Coenzyme a to Vanillin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4KKN
 
 | | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704 | | Descriptor: | Cytotoxic T-lymphocyte associated protein 4, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Bonanno, J, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Scott Glenn, A, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704
to be published
|
|
2H1P
 
 | | THE THREE-DIMENSIONAL STRUCTURES OF A POLYSACCHARIDE BINDING ANTIBODY TO CRYPTOCOCCUS NEOFORMANS AND ITS COMPLEX WITH A PEPTIDE FROM A PHAGE DISPLAY LIBRARY: IMPLICATIONS FOR THE IDENTIFICATION OF PEPTIDE MIMOTOPES | | Descriptor: | 2H1, PA1 | | Authors: | Young, A.C.M, Valadon, P, Casadevall, A, Scharff, M.D, Sacchettini, J.C. | | Deposit date: | 1997-11-12 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implications for the identification of peptide mimotopes.
J.Mol.Biol., 274, 1997
|
|
3RFZ
 
 | | Crystal structure of the FimD usher bound to its cognate FimC:FimH substrate | | Descriptor: | Chaperone protein fimC, Outer membrane usher protein, type 1 fimbrial synthesis, ... | | Authors: | Phan, G, Remaut, H, Lebedev, A, Geibel, S, Waksman, G. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
4N8P
 
 | | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Uncharacterized protein | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711]
to be published
|
|
4N5U
 
 | | Crystal structure of the 4th FN3 domain of human Protein Tyrosine phosphatase, receptor type F [PSI-NYSGRC-006240] | | Descriptor: | Receptor-type tyrosine-protein phosphatase F, SULFATE ION | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.456 Å) | | Cite: | Crystal structure of the 4th FN3 domain of human PTP, receptor F [PSI-NYSGRC-006240]
to be published
|
|
4NOF
 
 | | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Polymeric immunoglobulin receptor | | Authors: | Sampathkumar, P, Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor
to be published
|
|
4N68
 
 | | Crystal structure of an internal FN3 domain from human Contactin-5 [PSI-NYSGRC-005804] | | Descriptor: | Contactin-5, SULFATE ION | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an internal FN3 domain from human Contactin-5 [PSI-NYSGRC-005804]
to be published
|
|
4NOB
 
 | | Crystal structure of the 1st Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of the 1st Ig domain from mouse Polymeric Immunoglobulin receptor
to be published
|
|
4P5N
 
 | | Structure of CNAG_02591 from Cryptococcus neoformans | | Descriptor: | CADMIUM ION, GLYCEROL, Hypothetical protein CNAG_02591, ... | | Authors: | Ramagopal, U.A, McClelland, E.E, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A Small Protein Associated with Fungal Energy Metabolism Affects the Virulence of Cryptococcus neoformans in Mammals.
Plos Pathog., 12, 2016
|
|
4Q0C
 
 | | 3.1 A resolution crystal structure of the B. pertussis BvgS periplasmic domain | | Descriptor: | Virulence sensor protein BvgS | | Authors: | Dupre, E, Herrou, J, Lensink, M.F, Wintjens, R, Lebedev, A, Crosson, S, Villeret, V, Locht, C, Antoine, R, Jacob-Dubuisson, F. | | Deposit date: | 2014-04-01 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS.
Plos Pathog., 11, 2015
|
|
4HDI
 
 | | Crystal Structure of 3E5 IgG3 FAB from mus musculus | | Descriptor: | Ig heavy chain V region RF, Ig gamma-3 chain C region, Kappa light chain variable region, ... | | Authors: | Janda, A, Eryilmaz, E, Kim, J, Cordero, R.J.B, Cowburn, D, Casadevall, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Global structures of IgG isotypes expressing identical variable regions.
Mol.Immunol., 56, 2013
|
|
4P9G
 
 | | Structure of the 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. | | Descriptor: | 2,4'-dihydroxyacetophenone dioxygenase, CARBONATE ION, FE (III) ION, ... | | Authors: | Keegan, R, Lebedev, A, Erskine, P, Guo, J, Wood, S.P, Hopper, D.J, Cooper, J.B. | | Deposit date: | 2014-04-03 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. 4HAP
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1S31
 
 | | Crystal Structure Analysis of the human Tub protein (isoform a) spanning residues 289 through 561 | | Descriptor: | TRIETHYLENE GLYCOL, tubby isoform a | | Authors: | Boutboul, S, Carroll, K.J, Basdevant, A, Gomez, C, Nandrot, E, Clement, K, Shapiro, L, Abitbol, M. | | Deposit date: | 2004-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | A novel human obesity and sensory deficit syndrome resulting from a mutation in the TUB gene
To be Published
|
|
5T68
 
 | | Crystal structure of Syk catalytic domain in complex with a furo[3,2-d]pyrimidine | | Descriptor: | N~4~-cyclopropyl-N~2~-(3-methyl-1H-indazol-6-yl)furo[3,2-d]pyrimidine-2,4-diamine, Tyrosine-protein kinase SYK | | Authors: | Argiriadi, M.A, Hoemann, M, Wilson, N, Banach, D, Burchat, A, Calderwood, D, Clapham, B, Cox, P, Duignan, D.B, Konopacki, D, Somal, G, Vasudevan, A. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Synthesis and optimization of furano[3,2-d]pyrimidines as selective spleen tyrosine kinase (Syk) inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
6LK2
 
 | | Crystal structure of Providencia alcalifaciens 3-dehydroquinate synthase (DHQS) in complex with Mg2+, NAD and chlorogenic acid | | Descriptor: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 3-dehydroquinate synthase, ... | | Authors: | Neetu, N, Katiki, M, Kumar, P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural and Biochemical Analyses Reveal that Chlorogenic Acid Inhibits the Shikimate Pathway.
J.Bacteriol., 202, 2020
|
|
6LLA
 
 | | Crystal structure of Providencia alcalifaciens 3-dehydroquinate synthase (DHQS) in complex with Mg2+ and NAD | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Neetu, N, Katiki, M, Kumar, P. | | Deposit date: | 2019-12-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Biochemical Analyses Reveal that Chlorogenic Acid Inhibits the Shikimate Pathway.
J.Bacteriol., 202, 2020
|
|
4B15
 
 | | crystal structure of tamarind chitinase like lectin (TCLL) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Patil, D.N, Kumar, P. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Investigation of a Novel N-Acetyl Glucosamine Binding Chi-Lectin which Reveals Evolutionary Relationship with Class III Chitinases.
Plos One, 8, 2013
|
|
