6PVE
 
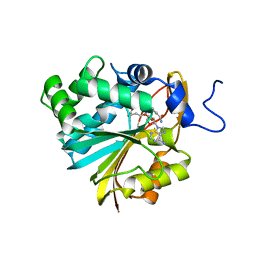 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL319 | | Descriptor: | 9-(5-{[(3S)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)propyl]amino}-5-deoxy-alpha-D-ribofuranosyl)-9H-purin-6-amine, NNMT protein | | Authors: | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | Deposit date: | 2019-07-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
5ZNC
 
 | |
5ZNI
 
 | | Plasmodium falciparum purine nucleoside phosphorylase in complex with mefloquine | | Descriptor: | (11R,12S)- Mefloquine, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Chen, D, Nordlund, P, Dziekan, J.M. | | Deposit date: | 2018-04-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identifying purine nucleoside phosphorylase as the target of quinine using cellular thermal shift assay.
Sci Transl Med, 11, 2019
|
|
5TUC
 
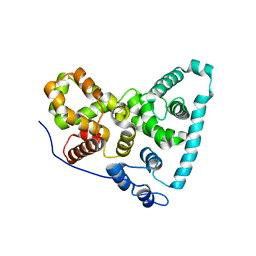 | | Crystal Structure of the Sus TBC1D15 GAP Domain | | Descriptor: | Sus TBC1D15 GAP Domain | | Authors: | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
5TUB
 
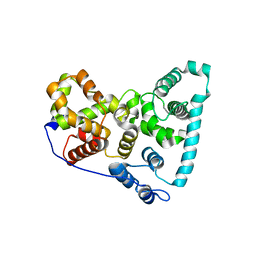 | | Crystal Structure of the Shark TBC1D15 GAP Domain | | Descriptor: | Shark TBC1D15 GTPase-activating Protein | | Authors: | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
4G1T
 
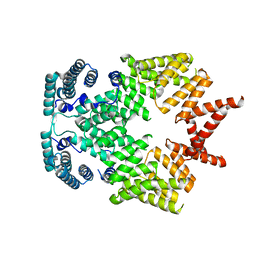 | | Crystal structure of interferon-stimulated gene 54 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 2 | | Authors: | Yang, Z, Liang, H, Zhou, Q, Li, Y, Chen, H, Ye, W, Chen, D, Fleming, J, Shu, H, Liu, Y. | | Deposit date: | 2012-07-11 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms.
Cell Res., 22, 2012
|
|
1XAV
 
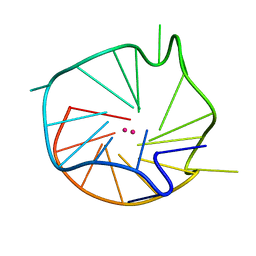 | | Major G-quadruplex structure formed in human c-MYC promoter, a monomeric parallel-stranded quadruplex | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3', POTASSIUM ION | | Authors: | Ambrus, A, Chen, D, Dai, J, Jones, R.A, Yang, D. | | Deposit date: | 2004-08-26 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the biologically relevant G-Quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization.
Biochemistry, 44, 2005
|
|
2F8U
 
 | | G-quadruplex structure formed in human Bcl-2 promoter, hybrid form | | Descriptor: | 5'-D(*GP*GP*GP*CP*GP*CP*GP*GP*GP*AP*GP*GP*AP*AP*TP*TP*GP*GP*GP*CP*GP*GP*G)-3' | | Authors: | Dai, J, Chen, D, Carver, M, Yang, D. | | Deposit date: | 2005-12-03 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region.
Nucleic Acids Res., 34, 2006
|
|
6WH8
 
 | | The structure of NTMT1 in complex with compound BM-30 | | Descriptor: | 4HP-PRO-LYS-ARG-NH2, BM-30, N-terminal Xaa-Pro-Lys N-methyltransferase 1, ... | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Selective Peptidomimetic Inhibitors of NTMT1/2: Rational Design, Synthesis, Characterization, and Crystallographic Studies.
J.Med.Chem., 63, 2020
|
|
6XZ1
 
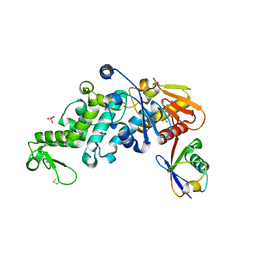 | | Conjugate of the HECT domain of HUWE1 with ubiquitin | | Descriptor: | HECT, UBA and WWE domain containing 1, isoform CRA_a, ... | | Authors: | Liu, B, Seenivasan, A, Nair, R, Chen, D, Lowe, E.D, Lorenz, S. | | Deposit date: | 2020-01-31 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution and Structural Analysis of a HECT Ligase-Ubiquitin Complex via an Activity-Based Probe.
Acs Chem.Biol., 16, 2021
|
|
4BUL
 
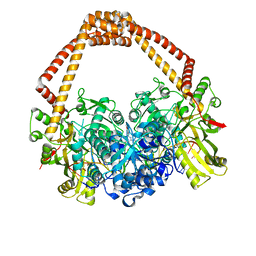 | | Novel hydroxyl tricyclics (e.g. GSK966587) as potent inhibitors of bacterial type IIA topoisomerases | | Descriptor: | (S)-4-((4-(((2,3-dihydro-[1,4]dioxino[2,3-c]pyridin-7-yl)methyl)amino)piperidin-1-yl)methyl)-3-fluoro-4-hydroxy-4H-pyrrolo[3,2,1-de][1,5]naphthyridin-7(5H)-one, 5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*AP*CP *CP*GP*CP*AP*C)-3', 5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*TP*AP*CP*CP*TP *AP*CP*GP*GP*CP*T)-3', ... | | Authors: | Miles, T.J, Hennessy, A.J, Bax, B, Brooks, G, Brown, B.S, Brown, P, Cailleau, N, Chen, D, Dabbs, S, Davies, D.T, Esken, J.M, Giordano, I, Hoover, J.L, Huang, J, Jones, G.E, Sukmar, S.K.K, Spitzfaden, C, Markwell, R.E, Minthorn, E.A, Rittenhouse, S, Gwynn, M.N, Pearson, N.D. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Hydroxyl Tricyclics (E.G., Gsk966587) as Potent Inhibitors of Bacterial Type Iia Topoisomerases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
4IMO
 
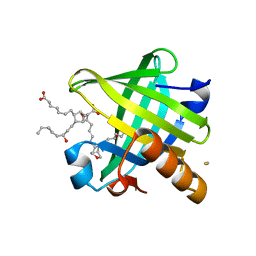 | | Crystal structure of wild type human Lipocalin PGDS in complex with substrate analog U44069 | | Descriptor: | (5E)-7-{(1R,4S,5S,6R)-5-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-6-yl}hept-5-enoic acid, Lipocalin-type prostaglandin-D synthase, THIOCYANATE ION | | Authors: | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | Deposit date: | 2013-01-03 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
4JDN
 
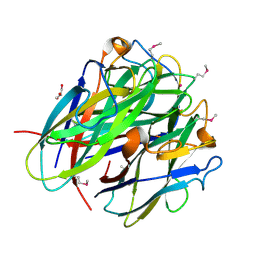 | | Secreted Chlamydial Protein PGP3, C-terminal Domain | | Descriptor: | GLYCEROL, POTASSIUM ION, Virulence plasmid protein pGP3-D | | Authors: | Galaleldeen, A, Taylor, A.B, Chen, D, Holloway, S.P, Zhong, G, Hart, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Chlamydia trachomatis immunodominant antigen Pgp3.
J.Biol.Chem., 288, 2013
|
|
4JDO
 
 | | Secreted chlamydial protein pgp3, coiled-coil deletion | | Descriptor: | SODIUM ION, Virulence plasmid protein pGP3-D | | Authors: | Galaleldeen, A, Taylor, A.B, Chen, D, Holloway, S.P, Zhong, G, Hart, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Chlamydia trachomatis immunodominant antigen Pgp3.
J.Biol.Chem., 288, 2013
|
|
4JDM
 
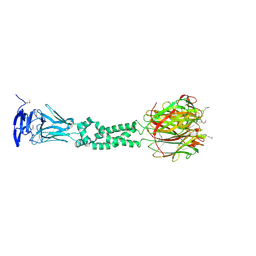 | | Secreted Chlamydial Protein PGP3, full-length | | Descriptor: | Virulence plasmid protein pGP3-D | | Authors: | Galaleldeen, A, Taylor, A.B, Chen, D, Holloway, S.P, Zhong, G, Hart, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Chlamydia trachomatis immunodominant antigen Pgp3.
J.Biol.Chem., 288, 2013
|
|
4IMN
 
 | | Crystal structure of wild type human Lipocalin PGDS bound with PEG MME 2000 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lipocalin-type prostaglandin-D synthase | | Authors: | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | Deposit date: | 2013-01-03 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
6AHZ
 
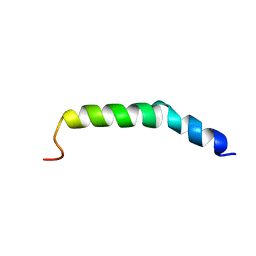 | | The NMR Structure of the Polysialyltranseferase Domain (PSTD) in Polysialyltransferase ST8siaIV | | Descriptor: | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase | | Authors: | Liu, X.H, Lu, B, Peng, L.X, Liao, S.M, Zhou, F, Chen, D, Lu, Z.L, Zhou, G.P, Huang, R.B. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Inhibition of Polysialyltranseferase ST8SiaIV Through Heparin Binding to Polysialyltransferase Domain (PSTD).
Med Chem, 15, 2019
|
|
9CUO
 
 | | Crystal structure of CRBN with compound 3 | | Descriptor: | (3S)-3-(3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, Protein cereblon, ... | | Authors: | Zheng, X, Ji, N, Campbell, V, Slavin, A, Zhu, X, Chen, D, Rong, H, Enerson, B, Mayo, M, Sharma, K, Browne, C.M, Klaus, C.R, Li, H, Massa, G, McDonald, A.A, Shi, Y, Sintchak, M, Skouras, S, Walther, D.M, Yuan, K, Zhang, Y, Kelleher, J, Guang, L, Luo, X, Mainolfi, N, Weiss, M.M. | | Deposit date: | 2024-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
J.Med.Chem., 67, 2024
|
|
9D0V
 
 | | Crystal structure of CDK2/CyclinE1 in complex with Cpd 2 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-8-cyclopentyl-2-[4-(ethanesulfonyl)-2-methylanilino]pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, ... | | Authors: | Kwiatkowski, N, Liang, T, Sha, Z, Collier, P.N, Yang, A, Sathappa, M, Paul, A, Su, L, Zheng, X, Aversa, R, Li, K, Mehovic, R, Breitkopf, S.B, Chen, D, Howarth, C.L, Yuan, K, Jo, H, Growney, J.D, Weiss, M, Williams, J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | CDK2 heterobifunctional degraders co-degrade CDK2 and cyclin E resulting in efficacy in CCNE1-amplified and overexpressed cancers.
Cell Chem Biol, 32, 2025
|
|
9D0W
 
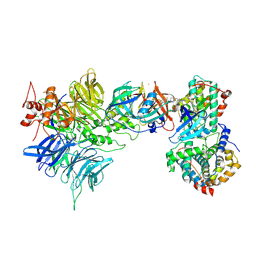 | | Cryo-EM structure of CDK2/CyclinE1 in complex with CRBN/DDB1 and Cpd 4 | | Descriptor: | (3R)-3-(5-{4-[(2-{4-[(8-cyclopentyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl)amino]-3-methylbenzene-1-sulfonyl}-7-azaspiro[3.5]nonan-7-yl)methyl]piperidin-1-yl}-4-fluoro-3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, Cyclin-dependent kinase 2, DNA damage-binding protein 1, ... | | Authors: | Kwiatkowski, N, Liang, T, Sha, Z, Collier, P.N, Yang, A, Sathappa, M, Paul, A, Su, L, Zheng, X, Aversa, R, Li, K, Mehovic, R, Breitkopf, S.B, Chen, D, Howarth, C.L, Yuan, K, Jo, H, Growney, J.D, Weiss, M, Williams, J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | CDK2 heterobifunctional degraders co-degrade CDK2 and cyclin E resulting in efficacy in CCNE1-amplified and overexpressed cancers.
Cell Chem Biol, 32, 2025
|
|
9D0X
 
 | | Cryo-EM structure of CDK2/CyclinE1 in complex with CRBN/DDB1 and Cpd 4 (local mask) | | Descriptor: | (3R)-3-(5-{4-[(2-{4-[(8-cyclopentyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl)amino]-3-methylbenzene-1-sulfonyl}-7-azaspiro[3.5]nonan-7-yl)methyl]piperidin-1-yl}-4-fluoro-3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1, ... | | Authors: | Kwiatkowski, N, Liang, T, Sha, Z, Collier, P.N, Yang, A, Sathappa, M, Paul, A, Su, L, Zheng, X, Aversa, R, Li, K, Mehovic, R, Breitkopf, S.B, Chen, D, Howarth, C.L, Yuan, K, Jo, H, Growney, J.D, Weiss, M, Williams, J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | CDK2 heterobifunctional degraders co-degrade CDK2 and cyclin E resulting in efficacy in CCNE1-amplified and overexpressed cancers.
Cell Chem Biol, 32, 2025
|
|
