6C04
 
 | | Mtb RNAP Holo/RbpA/double fork DNA -closed clamp | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
3TBI
 
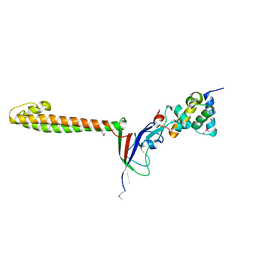 | |
5VI8
 
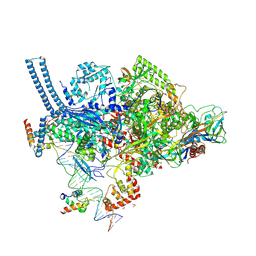 | | Structure of a mycobacterium smegmatis transcription initiation complex with an upstream-fork promoter fragment | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Hubin, E.A, Campbell, E.A, Darst, S.A. | | Deposit date: | 2017-04-14 | | Release date: | 2017-04-26 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures.
Nat Commun, 8, 2017
|
|
6BZO
 
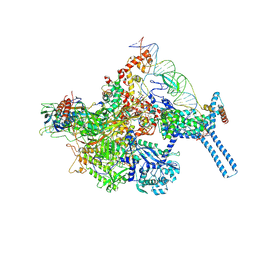 | | Mtb RNAP Holo/RbpA/Fidaxomicin/upstream fork DNA | | Descriptor: | DNA (26-MER), DNA (32-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2017-12-25 | | Release date: | 2018-03-28 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
7UO7
 
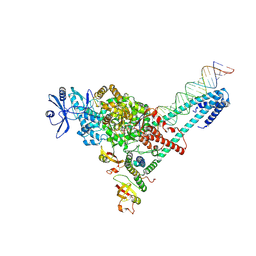 | | SARS-CoV-2 replication-transcription complex bound to ATP, in a pre-catalytic state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO4
 
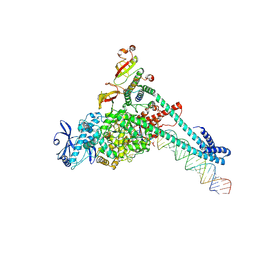 | | SARS-CoV-2 replication-transcription complex bound to Remdesivir triphosphate, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOB
 
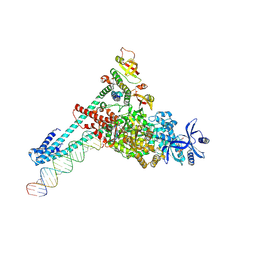 | | SARS-CoV-2 replication-transcription complex bound to GTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOE
 
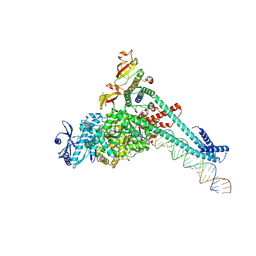 | | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO9
 
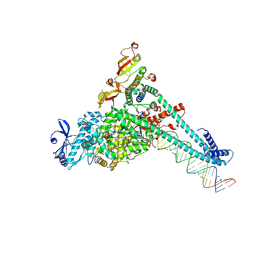 | | SARS-CoV-2 replication-transcription complex bound to UTP, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
5VI5
 
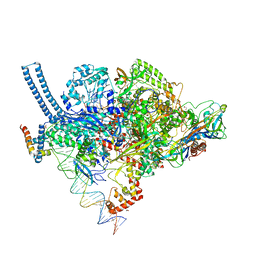 | | Structure of Mycobacterium smegmatis transcription initiation complex with a full transcription bubble | | Descriptor: | 1,2-ETHANEDIOL, DNA (44-MER), DNA (49-MER), ... | | Authors: | Darst, S.A, Campbell, E.A, Lilic, M. | | Deposit date: | 2017-04-14 | | Release date: | 2017-07-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures.
Nat Commun, 8, 2017
|
|
3GFK
 
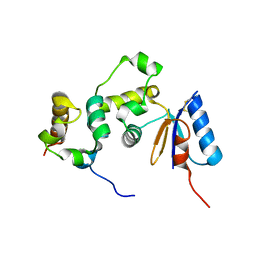 | | Crystal structure of Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, Regulatory protein spx | | Authors: | Lamour, V, Westblade, L.F, Campbell, E.A, Darst, S.A. | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex
J.Struct.Biol., 168, 2009
|
|
7L7B
 
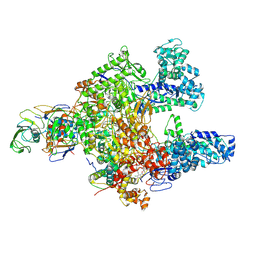 | | Clostridioides difficile RNAP with fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Boyaci, H, Campbell, E.A, Darst, S.A, Chen, J. | | Deposit date: | 2020-12-28 | | Release date: | 2022-02-02 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Basis of narrow-spectrum activity of fidaxomicin on Clostridioides difficile.
Nature, 604, 2022
|
|
9DJ8
 
 | |
4XAX
 
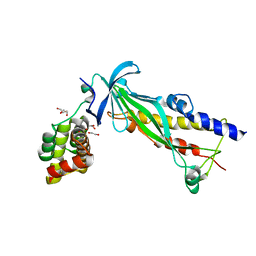 | | Crystal structure of Thermus thermophilus CarD in complex with the Thermus aquaticus RNA polymerase beta1 domain | | Descriptor: | 1,2-ETHANEDIOL, CarD, DNA-directed RNA polymerase subunit beta domain 1 | | Authors: | Chen, J, Bae, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex.
Elife, 4, 2015
|
|
8SQJ
 
 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQ9
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQK
 
 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8T02
 
 | | Reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA: unwinding duplex DNA (rPTCi) | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-31 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8T00
 
 | | Reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA: closed duplex DNA (rPTCc) | | Descriptor: | DNA (26-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-31 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8T0L
 
 | | E. coli Sw2/Snf2 ATPase RapA bound to both ADP-AlF3 and reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA (29-MER), ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-06-01 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8SZW
 
 | | Reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA: open duplex DNA (rPTCo) | | Descriptor: | DNA (25-MER), DNA (27-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-30 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7KRN
 
 | | Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KIF
 
 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with WhiB7 transcription factor | | Descriptor: | DNA (55-MER), DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7.
Mol.Cell, 81, 2021
|
|
7KIM
 
 | | Mycobacterium tuberculosis WT RNAP transcription closed promoter complex with WhiB7 transcription factor | | Descriptor: | DNA (45-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7.
Mol.Cell, 81, 2021
|
|
7KRP
 
 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - BTC (local refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPSO, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
