1H7K
 
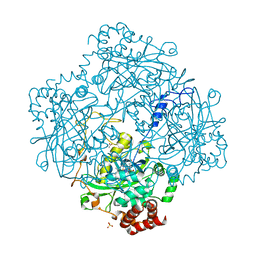 | | Formation of a tyrosyl radical intermediate in Proteus mirabilis catalase by directed mutagenesis and consequences for nucleotide reactivity | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Gambarelli, S, Gaillard, J, Sainz, G, Stojanoff, V, Jouve, H.M. | | Deposit date: | 2001-07-08 | | Release date: | 2004-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Formation of a Tyrosyl Radical Intermediate in Proteus Mirabilis Catalase by Directed Mutagenesis and Consequences for Nucleotide Reactivity.
Biochemistry, 40, 2001
|
|
1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
6HTO
 
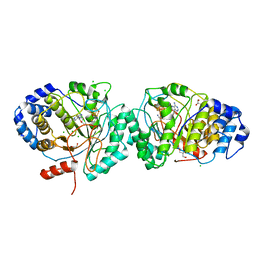 | | Tryptophan lyase 'empty state' | | Descriptor: | 3-methyl-2-indolic acid synthase, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | Authors: | Amara, P, Mouesca, J.M, Bella, M, Martin, L, Saragaglia, C, Gambarelli, S, Nicolet, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Radical S-Adenosyl-l-methionine Tryptophan Lyase (NosL): How the Protein Controls the Carboxyl Radical •CO2-Migration.
J.Am.Chem.Soc., 140, 2018
|
|
6HTM
 
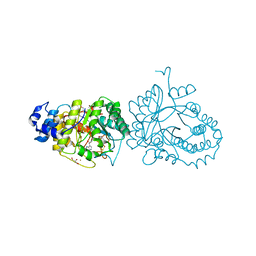 | | X-ray structure of the tryptophan lyase NosL in complex with bound tryptamin | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, 3-methyl-2-indolic acid synthase, 5'-DEOXYADENOSINE, ... | | Authors: | Amara, P, Mouesca, J.M, Bella, M, Saragaglia, C, Gambarelli, S, Nicolet, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Radical S-Adenosyl-l-methionine Tryptophan Lyase (NosL): How the Protein Controls the Carboxyl Radical •CO2-Migration.
J.Am.Chem.Soc., 140, 2018
|
|
6HTK
 
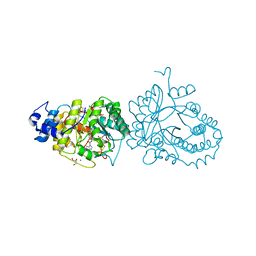 | | X-ray structure of the tryptophan lyase NosL in complex with (R)-(+)-indoline-2-carboxylate | | Descriptor: | 1H-indole-2-carboxylic acid, 3-methyl-2-indolic acid synthase, 5'-DEOXYADENOSINE, ... | | Authors: | Amara, P, Mouesca, J.M, Bella, M, Martin, L, Saragaglia, C, Gambarelli, S, Nicolet, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Radical S-Adenosyl-l-methionine Tryptophan Lyase (NosL): How the Protein Controls the Carboxyl Radical •CO2-Migration.
J.Am.Chem.Soc., 140, 2018
|
|
3EPV
 
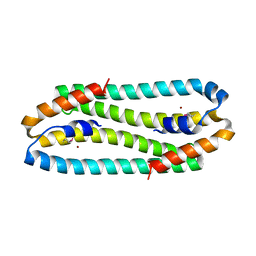 | | X-ray Structure of the Metal-sensor CnrX in both the Apo- and Copper-bound Forms | | Descriptor: | COPPER (II) ION, Nickel and cobalt resistance protein cnrR | | Authors: | Pompidor, G, Maillard, A.P, Girard, E, Gambarelli, S, Kahn, R, Coves, J. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | X-ray structure of the metal-sensor CnrX in both the apo- and copper-bound forms.
Febs Lett., 2008
|
|
4OKP
 
 | |
1CX5
 
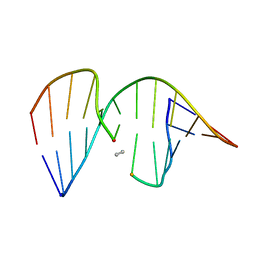 | | ANTISENSE DNA/RNA HYBRID CONTAINING MODIFIED BACKBONE | | Descriptor: | 5'-D(*CP*GP*CP*GP*TP*T*(MMT)P*TP*GP*CP*GP*C), 5'-R(*GP*CP*GP*CP*AP*AP*AP*AP*CP*GP*CP*G) | | Authors: | Yang, X, Han, X, Cross, C, Sanghvi, Y, Gao, X. | | Deposit date: | 1999-08-28 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of an antisense DNA.RNA hybrid duplex containing a 3'-CH(2)N(CH(3))-O-5' or an MMI backbone linker.
Biochemistry, 38, 1999
|
|
2OEY
 
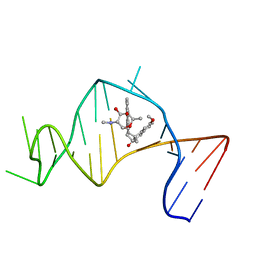 | | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA | | Descriptor: | (1R,3A'S,10'S,10A'R)-7-METHOXY-2-OXO-10',10A'-DIHYDRO-2H,3A'H-SPIRO[NAPHTHALENE-1,3'-PENTALENO[1,2-B]NAPHTHALEN]-10'-YL 2,6-DIDEOXY-2-(METHYLAMINO)-ALPHA-D-GALACTOPYRANOSIDE, DNA (25-MER) | | Authors: | Zhang, N, Lin, Y, Xiao, Z, Jones, G.B, Goldberg, I.H. | | Deposit date: | 2007-01-01 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA.
Biochemistry, 46, 2007
|
|
7K9Z
 
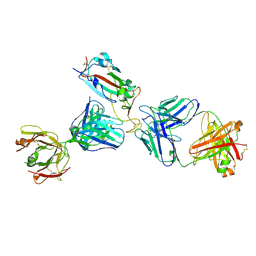 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with the Fab fragments of neutralizing antibodies 298 and 52 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 298 Fab Heavy Chain, 298 Fab Light Chain, ... | | Authors: | Newton, J.C, Kucharska, I, Rujas, E, Cui, H, Julien, J.P. | | Deposit date: | 2020-09-29 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers.
Nat Commun, 12, 2021
|
|
8DNN
 
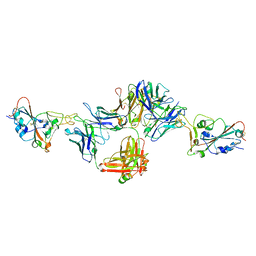 | | Crystal structure of neutralizing antibody 80 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 80 FAB HEAVY CHAIN, 80 FAB LIGHT CHAIN, ... | | Authors: | Muthuraman, K, Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A multi-specific, multi-affinity antibody platform neutralizes sarbecoviruses and confers protection against SARS-CoV-2 in vivo.
Sci Transl Med, 15, 2023
|
|
2Y3H
 
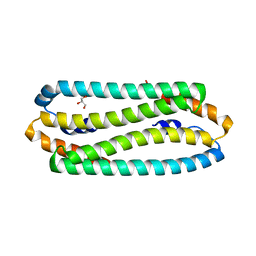 | | E63Q mutant of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | GLYCEROL, NICKEL AND COBALT RESISTANCE PROTEIN CNRR | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
2Y3B
 
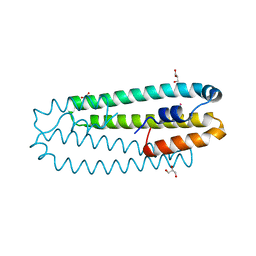 | | Co-bound form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | COBALT (II) ION, GLYCEROL, NICKEL AND COBALT RESISTANCE PROTEIN CNRR | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
2Z3S
 
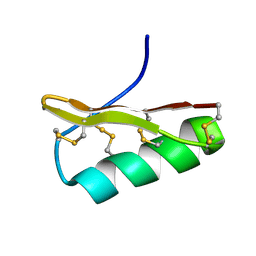 | | NMR structure of AgTx2-MTX | | Descriptor: | AgTx2-MTX | | Authors: | Pimentel, C, M'Barrek, S, Visan, V, Grissmer, S, Sabatier, J.M, Darbon, H, Fajloun, Z. | | Deposit date: | 2007-06-06 | | Release date: | 2008-04-22 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Chemical synthesis and 1H-NMR 3D structure determination of AgTx2-MTX chimera, a new potential blocker for Kv1.2 channel, derived from MTX and AgTx2 scorpion toxins.
Protein Sci., 17, 2008
|
|
1PJV
 
 | | Cobatoxin 1 from Centruroides noxius Scorpion venom: Chemical Synthesis, 3-D Structure in Solution, Pharmacology and Docking on K+ channels | | Descriptor: | Cobatoxin 1 | | Authors: | Mosbah, A, Jouirou, B, Visan, V, Grissmer, S, El Ayeb, M, Rochat, H, De Waard, M, Mabrouk, K, Sabatier, J.M. | | Deposit date: | 2003-06-03 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Cobatoxin 1 from Centruroides noxius scorpion venom: chemical synthesis, three-dimensional structure in solution, pharmacology and docking on K+ channels.
Biochem.J., 377, 2004
|
|
4JC0
 
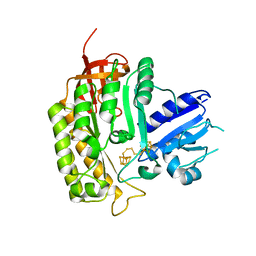 | | Crystal structure of Thermotoga maritima holo RimO in complex with pentasulfide, Northeast Structural Genomics Consortium Target VR77 | | Descriptor: | IRON/SULFUR PENTA-SULFIDE CONNECTED CLUSTERS, Ribosomal protein S12 methylthiotransferase RimO | | Authors: | Forouhar, F, Hussain, M, Seetharaman, J, Fang, Y, Chen, C.X, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases.
Nat.Chem.Biol., 9, 2013
|
|
6FBJ
 
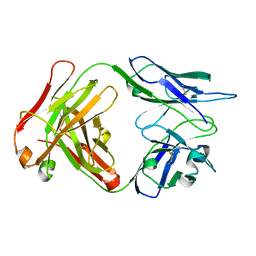 | | monoclonal antibody targeting Matrix metalloproteinase 7 | | Descriptor: | Heavy Chain, Light chain | | Authors: | Dym, O. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel monoclonal antibody targeting Matrix metalloproteinase 7 shows therapeutic potential against pancreatic cancer
To Be Published
|
|
4E3J
 
 | |
4E3I
 
 | |
4E3L
 
 | |
4E3K
 
 | |
4E3N
 
 | |
4E3M
 
 | |
4E3O
 
 | |
2WTP
 
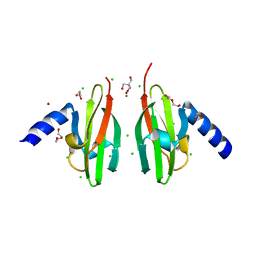 | | Crystal Structure of Cu-form Czce from C. metallidurans CH34 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Haertlein, I, Girard, E, Sarret, G, Hazemann, J, Gourhant, P, Kahn, R, Coves, J. | | Deposit date: | 2009-09-18 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evidence for Conformational Changes Upon Copper Binding to Cupriavidus Metallidurans Czce.
Biochemistry, 49, 2010
|
|
