7JX5
 
 | | Crystal Structure of N-Phenylalanine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Phenylalanine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices
J.Am.Chem.Soc., 143, 2021
|
|
2HTH
 
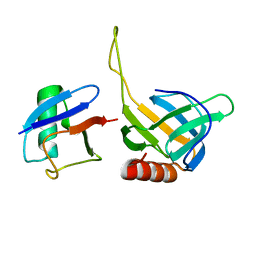 | | Structural basis for ubiquitin recognition by the human EAP45/ESCRT-II GLUE domain | | Descriptor: | Ubiquitin, Vacuolar protein sorting protein 36 | | Authors: | Alam, S.L, Whitby, F.G, Hill, C.P, Sundquist, W.I. | | Deposit date: | 2006-07-25 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
6E94
 
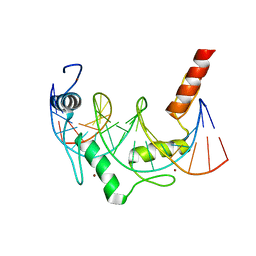 | | Crystal Structure of ZBTB38 C-terminal Zinc Fingers 6-9 K1055R in complex with methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*TP*CP*AP*TP*(DCM)P*GP*GP*(DCM)P*GP*CP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*(DCM)P*GP*CP*(DCM)P*GP*AP*TP*GP*AP*GP*TP*GP*C)-3'), ZINC ION, ... | | Authors: | Hudson, N.O, Whitby, F.G, Buck-Koehntop, B.A. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Structural insights into methylated DNA recognition by the C-terminal zinc fingers of the DNA reader protein ZBTB38.
J. Biol. Chem., 293, 2018
|
|
6E93
 
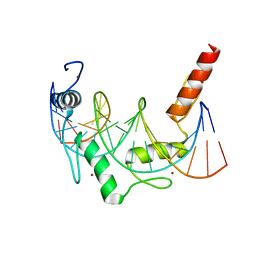 | | Crystal Structure of ZBTB38 C-terminal Zinc Fingers 6-9 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*TP*CP*AP*TP*(DCM)P*GP*GP*(DCM)P*GP*CP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*(DCM)P*GP*CP*(DCM)P*GP*AP*TP*GP*AP*GP*TP*GP*C)-3'), ZINC ION, ... | | Authors: | Hudson, N.O, Whitby, F.G, Buck-Koehntop, B.A. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structural insights into methylated DNA recognition by the C-terminal zinc fingers of the DNA reader protein ZBTB38.
J. Biol. Chem., 293, 2018
|
|
1YAU
 
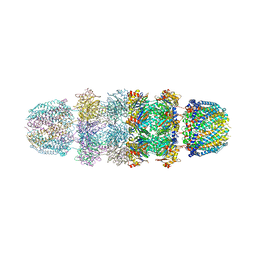 | | Structure of Archeabacterial 20S proteasome- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1YAR
 
 | | Structure of Archeabacterial 20S proteasome mutant D9S- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1YA7
 
 | | Implications for interactions of proteasome with PAN and PA700 from the 1.9 A structure of a proteasome-11S activator complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
4LCB
 
 | | Structure of Vps4 homolog from Acidianus hospitalis | | Descriptor: | CHLORIDE ION, Cell division protein CdvC, Vps4 | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-21 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4LGM
 
 | | Crystal Structure of Sulfolobus solfataricus Vps4 | | Descriptor: | CHLORIDE ION, Vps4 AAA ATPase | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
1Z7Q
 
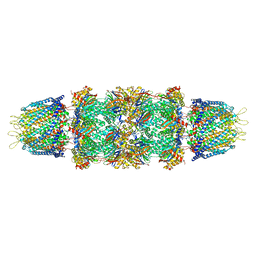 | | Crystal structure of the 20s proteasome from yeast in complex with the proteasome activator PA26 from Trypanosome brucei at 3.2 angstroms resolution | | Descriptor: | Potential proteasome component C5, Proteasome component C1, Proteasome component C11, ... | | Authors: | Forster, A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2005-03-26 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1JPH
 
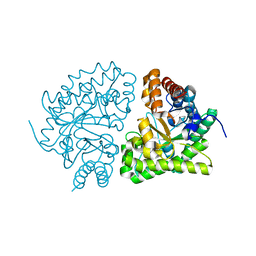 | | Ile260Thr mutant of Human UroD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
1JPI
 
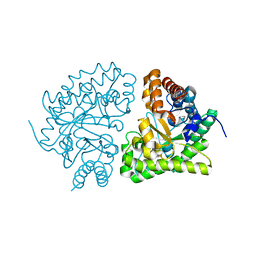 | | Phe232Leu mutant of human UROD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
1JPK
 
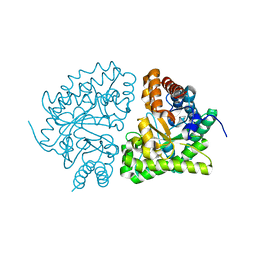 | | Gly156Asp mutant of Human UroD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
4WLR
 
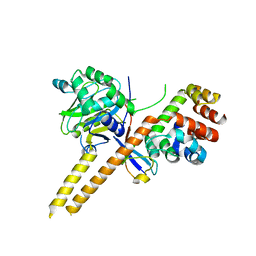 | | Crystal Structure of mUCH37-hRPN13 CTD-hUb complex | | Descriptor: | Polyubiquitin-B, Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
4WLP
 
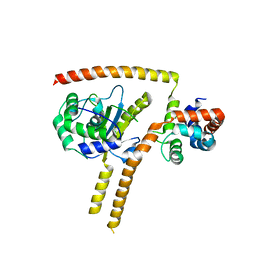 | | Crystal structure of UCH37-NFRKB Inhibited Deubiquitylating Complex | | Descriptor: | Nuclear factor related to kappa-B-binding protein, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
4WNN
 
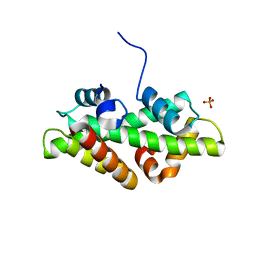 | | SPT16-H2A-H2B FACT HISTONE Complex | | Descriptor: | Histone H2A.1, Histone H2B.1, PHOSPHATE ION, ... | | Authors: | Kemble, D.J, Hill, C.P, Whitby, F.G, Formosa, T, McCullough, L.L. | | Deposit date: | 2014-10-13 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs.
Mol.Cell, 60, 2015
|
|
4V7O
 
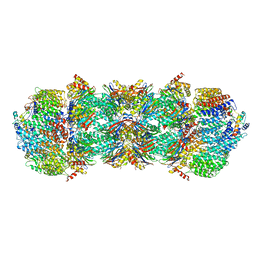 | | Proteasome Activator Complex | | Descriptor: | Proteasome activator BLM10, Proteasome component C1, Proteasome component C11, ... | | Authors: | Hill, C.P, Whitby, F.G. | | Deposit date: | 2009-12-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening.
Mol.Cell, 37, 2010
|
|
4WLQ
 
 | | Crystal structure of mUCH37-hRPN13 CTD complex | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
3OYX
 
 | | Haloferax volcanii Malate Synthase Magnesium/Glyoxylate Complex | | Descriptor: | CHLORIDE ION, GLYOXYLIC ACID, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C, Neighbor, A, Thomas, G, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-09-24 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
3PUG
 
 | | Haloferax volcanii Malate Synthase Native at 3mM Glyoxylate | | Descriptor: | CHLORIDE ION, GLYOXYLIC ACID, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C.D, Neighbor, A.M, Thomas, G.C, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-12-04 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
3OYZ
 
 | | Haloferax volcanii Malate Synthase Pyruvate/Acetyl-CoA Ternary Complex | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C, Neighbor, A, Thomas, G, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-09-24 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
3RBQ
 
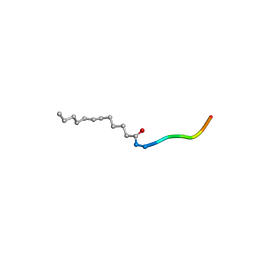 | | Co-crystal structure of human UNC119 (retina gene 4) and an N-terminal Transducin-alpha mimicking peptide | | Descriptor: | Guanine nucleotide-binding protein G(t) subunit alpha-1, Protein unc-119 homolog A | | Authors: | Constantine, R, Whitby, F.G, Hill, C.P, Baehr, W. | | Deposit date: | 2011-03-29 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | UNC119 is required for G protein trafficking in sensory neurons.
Nat.Neurosci., 14, 2011
|
|
1JR2
 
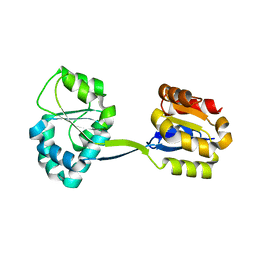 | | Structure of Uroporphyrinogen III Synthase | | Descriptor: | UROPORPHYRINOGEN-III SYNTHASE | | Authors: | Mathews, M.A, Schubert, H.L, Whitby, F.G, Alexander, K.J, Schadick, K, Bergonia, H.A, Phillips, J.D, Hill, C.P. | | Deposit date: | 2001-08-10 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of human uroporphyrinogen III synthase.
EMBO J., 20, 2001
|
|
3L37
 
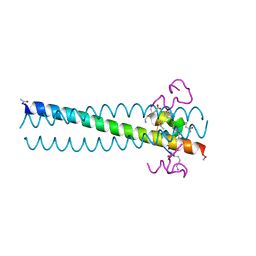 | | PIE12 D-peptide against HIV entry | | Descriptor: | GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
3L36
 
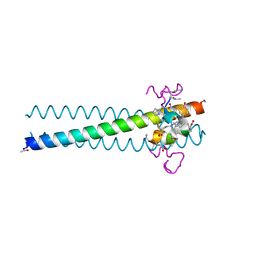 | | PIE12 D-peptide against HIV entry | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
