1D4B
 
 | | CIDE-N DOMAIN OF HUMAN CIDE-B | | Descriptor: | HUMAN CELL DEATH-INDUCING EFFECTOR B | | Authors: | Lugovskoy, A, Zhou, P, Chou, J, McCarty, J, Li, P, Wagner, G. | | Deposit date: | 1999-10-02 | | Release date: | 1999-12-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CLH
 
 | |
1DEC
 
 | |
1DRO
 
 | | NMR STRUCTURE OF THE CYTOSKELETON/SIGNAL TRANSDUCTION PROTEIN | | Descriptor: | BETA-SPECTRIN | | Authors: | Zhang, P, Talluri, S, Deng, H, Branton, D, Wagner, G. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pleckstrin homology domain of Drosophila beta-spectrin.
Structure, 3, 1995
|
|
5IJ4
 
 | | Solution structure of AN1-type zinc finger domain from Cuz1 (Cdc48 associated ubiquitin-like/zinc-finger protein-1) | | Descriptor: | CDC48-associated ubiquitin-like/zinc finger protein 1, ZINC ION | | Authors: | Sun, Z.-Y.J, Hanna, J, Wagner, G, Bhanu, M.K, Allan, M, Arthanari, H. | | Deposit date: | 2016-03-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cuz1 AN1 Zinc Finger Domain: An Exposed LDFLP Motif Defines a Subfamily of AN1 Proteins.
Plos One, 11, 2016
|
|
5A3G
 
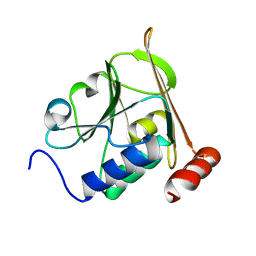 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A4H
 
 | | Solution structure of the lipid droplet anchoring peptide of CGI-58 bound to DPC micelles | | Descriptor: | 1-ACYLGLYCEROL-3-PHOSPHATE O-ACYLTRANSFERASE ABHD5 | | Authors: | Boeszoermenyi, A, Arthanari, H, Wagner, G, Nagy, H.M, Zangger, K, Lindermuth, H, Oberer, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Cgi-58 Motif Provides the Molecular Basis of Lipid Droplet Anchoring.
J.Biol.Chem., 290, 2015
|
|
5BXV
 
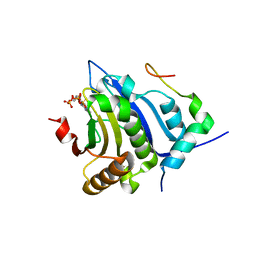 | | eIF4E complex | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Sekiyama, N, Arthanari, H, Papdopoulos, E, Rodriguez-Mias, R.A, Wagner, G, Leger-Abraham, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1Z9E
 
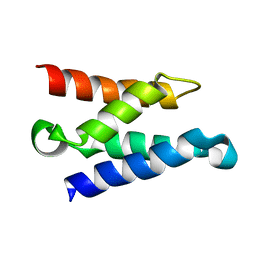 | | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75 | | Descriptor: | PC4 and SFRS1 interacting protein 2 | | Authors: | Cherepanov, P, Sun, Z.-Y.J, Rahman, S, Maertens, G, Wagner, G, Engelman, A. | | Deposit date: | 2005-04-01 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75
Nat.Struct.Mol.Biol., 12, 2005
|
|
6TIR
 
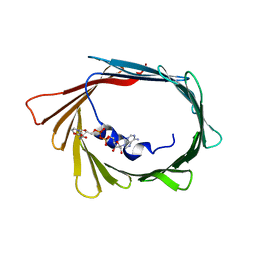 | |
6TIQ
 
 | |
1KRI
 
 | |
5JS7
 
 | | Structural model of a apo G-protein alpha subunit determined with NMR residual dipolar couplings and SAXS | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JS8
 
 | | Structural Model of a Protein alpha subunit in complex with GDP obtained with SAXS and NMR residual couplings | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7L0R
 
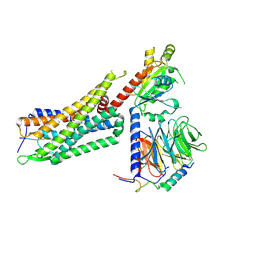 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0P
 
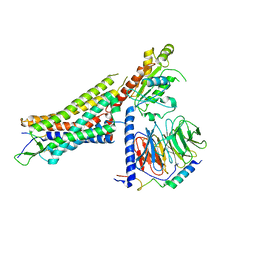 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
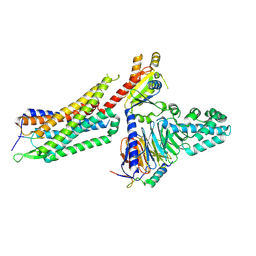 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4TQC
 
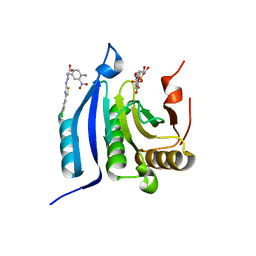 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2S)-3-(4-amino-3-nitrophenyl)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinyl}propanoic acid, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TPW
 
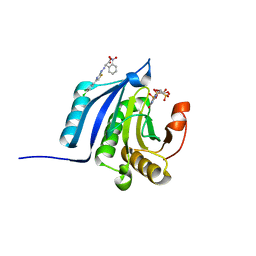 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TQB
 
 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(4-bromophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ME4
 
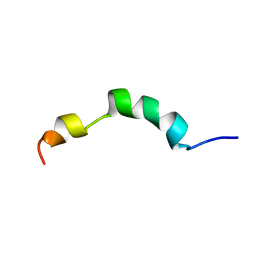 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME1
 
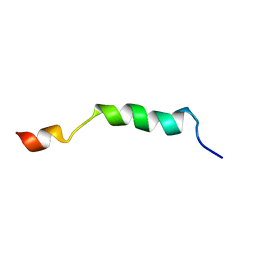 | | HIV-1 gp41 clade B double alanine mutant Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Gp41 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME2
 
 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME3
 
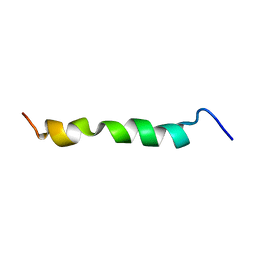 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
