3BUG
 
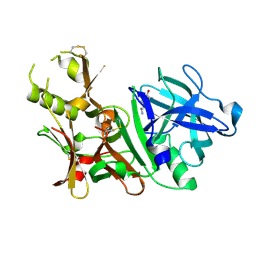 | | BACE-1 complexed with compound 3 | | Descriptor: | 4-(2-aminoethyl)-2-ethylphenol, BETA-SECRETASE 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7PQT
 
 | | Apo human Kv3.1 cryo-EM structure | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7PQU
 
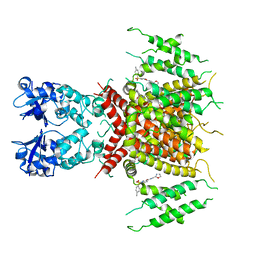 | | Ligand-bound human Kv3.1 cryo-EM structure (Lu AG00563) | | Descriptor: | 1-(4-methylphenyl)sulfonyl-N-(1,3-oxazol-2-ylmethyl)pyrrole-3-carboxamide, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
6DXM
 
 | |
6DVT
 
 | |
7OMT
 
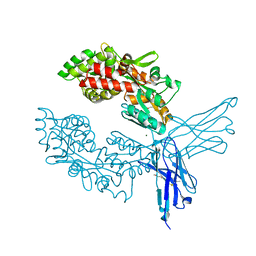 | | Crystal structure of ProMacrobody 21 with bound maltose | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7OMM
 
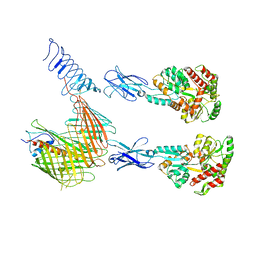 | | Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies (MBPs have not been built de novo) | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ProMacrobody 21,Maltodextrin-binding protein, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
2FYO
 
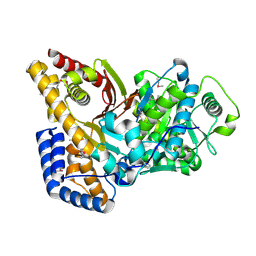 | | Crystal structure of rat carnitine palmitoyltransferase 2 in space group P43212 | | Descriptor: | Carnitine O-palmitoyltransferase II, mitochondrial | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
2FW3
 
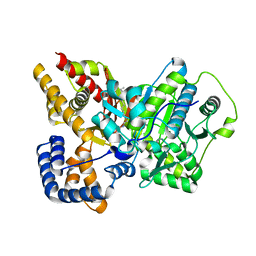 | | Crystal structure of rat carnitine palmitoyltransferase 2 in complex with antidiabetic drug ST1326 | | Descriptor: | (3R)-3-{[(TETRADECYLAMINO)CARBONYL]AMINO}-4-(TRIMETHYLAMMONIO)BUTANOATE, Carnitine O-palmitoyltransferase II, mitochondrial | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-01 | | Release date: | 2007-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
6DM7
 
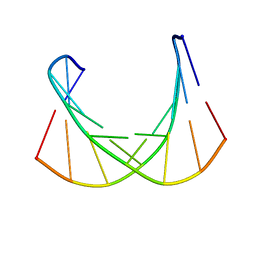 | |
6ED9
 
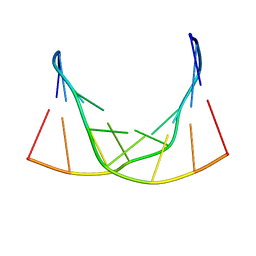 | |
3ODI
 
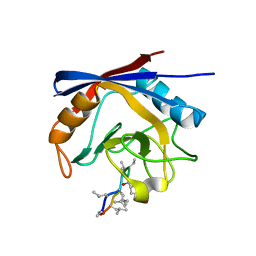 | | Crystal structure of cyclophilin A in complex with Voclosporin E-ISA247 | | Descriptor: | Cyclophilin A, Voclosporin | | Authors: | Kuglstatter, A, Stihle, M, Benz, J, Hennig, M. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ODL
 
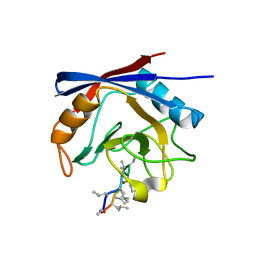 | | Crystal structure of cyclophilin A in complex with Voclosporin Z-ISA247 | | Descriptor: | Cyclophilin A, Voclosporin | | Authors: | Kuglstatter, A, Stihle, M, Benz, J, Hennig, M. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3MNO
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by (A611V, F608S) mutations at 1.55A | | Descriptor: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
3MNP
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by (A611V, V708A, E711G) mutations at 1.50A | | Descriptor: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
3MNE
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by F608S mutation at 1.96A | | Descriptor: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
7AA5
 
 | | Human TRPV4 structure in presence of 4a-PDD | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 4,Green fluorescent protein | | Authors: | Botte, M, Ulrich, A.K.G, Adaixo, R, Gnutt, D, Brockmann, A, Bucher, D, Chami, M, Bocquet, M, Ebbinghaus-Kintscher, U, Puetter, V, Becker, A, Egner, U, Stahlberg, H, Hennig, M, Holton, S.J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structural studies of the agonist complexed human TRPV4 ion-channel reveals novel structural rearrangements resulting in an open-conformation
To Be Published
|
|
3BUH
 
 | | BACE-1 complexed with compound 4 | | Descriptor: | 4-(2-aminoethyl)-2-cyclohexylphenol, beta-secretase 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1W6K
 
 | | Structure of human OSC in complex with Lanosterol | | Descriptor: | LANOSTEROL, LANOSTEROL SYNTHASE, octyl beta-D-glucopyranoside | | Authors: | Thoma, R, Schulz-Gasch, T, D'Arcy, B, Benz, J, Aebi, J, Dehmlow, H, Hennig, M, Ruf, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insight Into Steroid Scaffold Formation from the Structure of Human Oxidosqualene Cyclase
Nature, 432, 2004
|
|
1W6J
 
 | | Structure of human OSC in complex with Ro 48-8071 | | Descriptor: | LANOSTEROL SYNTHASE, TETRADECANE, [4-({6-[ALLYL(METHYL)AMINO]HEXYL}OXY)-2-FLUOROPHENYL](4-BROMOPHENYL)METHANONE, ... | | Authors: | Thoma, R, Schulz-Gasch, T, D'Arcy, B, Benz, J, Aebi, J, Dehmlow, H, Hennig, M, Ruf, A. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insight Into Steroid Scaffold Formation from the Structure of Human Oxidosqualene Cyclase
Nature, 432, 2004
|
|
1JUK
 
 | | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS IN A TRIGONAL CRYSTAL FORM | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Knoechel, T.R, Hennig, M, Merz, A, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1996-05-03 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of indole-3-glycerol phosphate synthase from the hyperthermophilic archaeon Sulfolobus solfataricus in three different crystal forms: effects of ionic strength.
J.Mol.Biol., 262, 1996
|
|
1JUL
 
 | | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS IN A SECOND ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Knoechel, T.R, Hennig, M, Merz, A, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1996-05-03 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of indole-3-glycerol phosphate synthase from the hyperthermophilic archaeon Sulfolobus solfataricus in three different crystal forms: effects of ionic strength.
J.Mol.Biol., 262, 1996
|
|
2DEB
 
 | | Crystal structure of rat carnitine palmitoyltransferase 2 in space group C2221 | | Descriptor: | COENZYME A, Carnitine O-palmitoyltransferase II, mitochondrial, ... | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-10 | | Release date: | 2007-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
4EP9
 
 | | CRYSTAL STRUCTURE OF RAT CARNITINE PALMITOYLTRANSFERASE 2 IN COMPLEX WITH CoA-site inhibitor | | Descriptor: | 4-[({1-[(5-chloro-2-methoxyphenyl)sulfonyl]-4-methyl-2,3-dihydro-1H-indol-6-yl}carbonyl)amino]benzoic acid, Carnitine O-palmitoyltransferase 2, mitochondrial, ... | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2012-04-17 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein.
FEBS Open Bio, 3, 2013
|
|
4EPH
 
 | | CRYSTAL STRUCTURE OF RAT CARNITINE PALMITOYLTRANSFERASE 2 IN COMPLEX with CoA-site inhibitor | | Descriptor: | 2-chloro-4-[({1-[(5-chloro-2-methoxyphenyl)sulfonyl]-4-methyl-2,3-dihydro-1H-indol-6-yl}carbonyl)amino]benzoic acid, Carnitine O-palmitoyltransferase 2, mitochondrial, ... | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2012-04-17 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein.
FEBS Open Bio, 3, 2013
|
|
