5ANY
 
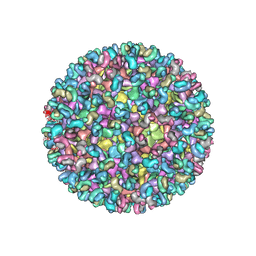 | | Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265 | | Descriptor: | E1, E2, FAB, ... | | Authors: | Fox, J.M, Long, F, Edeling, M.A, Lin, H, Duijl-Richter, M, Fong, R.H, Kahle, K.M, Smit, J.M, Jin, J, Simmons, G, Doranz, B.J, Crowe, J.E, Fremont, D.H, Rossmann, M.G, Diamond, M.S. | | Deposit date: | 2015-09-08 | | Release date: | 2015-11-25 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (16.9 Å) | | Cite: | Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress.
Cell(Cambridge,Mass.), 163, 2015
|
|
5UHY
 
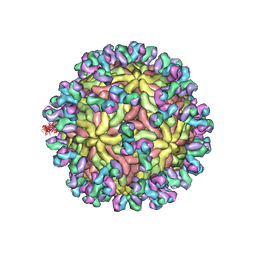 | | A Human Antibody Against Zika Virus Crosslinks the E Protein to Prevent Infection | | Descriptor: | ZV67 Fab chain 1, ZV67 Fab chain 2, envelope protein | | Authors: | Hasan, S.S, Miller, A, Sapparapu, G, Fernandez, E, Klose, T, Long, F, Fokine, A, Porta, J.C, Jiang, W, Diamond, M.S, Crowe Jr, J.E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2017-01-12 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A human antibody against Zika virus crosslinks the E protein to prevent infection.
Nat Commun, 8, 2017
|
|
8EWF
 
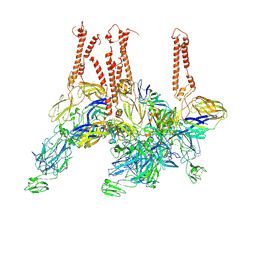 | |
8FXC
 
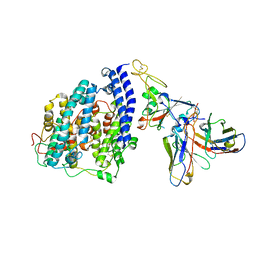 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8FXB
 
 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
7TE1
 
 | |
7SWX
 
 | |
7SWW
 
 | |
7BEM
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-269 Vh domain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEH
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-316 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEN
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253 and COVOX-75 Fabs | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEP
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-384 and S309 Fabs | | Descriptor: | CHLORIDE ION, COVOX-384 heavy chain, COVOX-384 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEO
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253H55L and COVOX-75 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEL
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-88 and COVOX-45 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COVOX-45 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEK
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEI
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-150 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEJ
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7TZ0
 
 | |
7TYZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DARPin FSR22, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
7UHC
 
 | |
7UHB
 
 | |
8SQN
 
 | | CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DuD1MoD2 chimeric MXRA8, E1 envelope glycoprotein, ... | | Authors: | Zimmerman, M.I, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Vertebrate-class-specific binding modes of the alphavirus receptor MXRA8.
Cell, 186, 2023
|
|
8UFA
 
 | |
8UFC
 
 | |
8UFB
 
 | |
