8RXB
 
 | |
1A0E
 
 | | XYLOSE ISOMERASE FROM THERMOTOGA NEAPOLITANA | | Descriptor: | COBALT (II) ION, XYLOSE ISOMERASE | | Authors: | Gallay, O, Chopra, R, Conti, E, Brick, P, Blow, D. | | Deposit date: | 1997-11-28 | | Release date: | 1998-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Class II Xylose Isomerases from Two Thermophiles and a Hyperthermophile
To be Published
|
|
3ICQ
 
 | | Karyopherin nuclear state | | Descriptor: | Exportin-T, GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cook, A.G, Fukuhara, N, Jinek, M, Conti, E. | | Deposit date: | 2009-07-18 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the tRNA export factor in the nuclear and cytosolic states
Nature, 461, 2009
|
|
3IBV
 
 | | Karyopherin cytosolic state | | Descriptor: | CALCIUM ION, Exportin-T | | Authors: | Cook, A.G, Fukuhara, N, Jinek, M, Conti, E. | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the tRNA export factor in the nuclear and cytosolic states
Nature, 461, 2009
|
|
3FHT
 
 | |
3FHC
 
 | | Crystal structure of human Dbp5 in complex with Nup214 | | Descriptor: | ATP-dependent RNA helicase DDX19B, Nuclear pore complex protein Nup214 | | Authors: | von Moeller, H, Conti, E. | | Deposit date: | 2008-12-09 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner
Nat.Struct.Mol.Biol., 16, 2009
|
|
4TXD
 
 | | Crystal structure of Thermofilum pendens Csc2 | | Descriptor: | Csc2, ZINC ION | | Authors: | Hrle, A, Conti, E. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family.
Rna Biol., 11, 2014
|
|
4U4C
 
 | | The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DOB1, CHLORIDE ION, ... | | Authors: | Falk, S, Weir, J.R, Hentschel, J, Reichelt, P, Bonneau, F, Conti, E. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Molecular Architecture of the TRAMP Complex Reveals the Organization and Interplay of Its Two Catalytic Activities.
Mol.Cell, 55, 2014
|
|
4W8W
 
 | |
4W8Z
 
 | |
2VXG
 
 | | Crystal structure of the conserved C-terminal region of Ge-1 | | Descriptor: | CG6181-PA, ISOFORM A | | Authors: | Jinek, M, Eulalio, A, Lingel, A, Helms, S, Conti, E, Izaurralde, E. | | Deposit date: | 2008-07-04 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-Terminal Region of Ge-1 Presents Conserved Structural Features Required for P-Body Localization.
RNA, 14, 2008
|
|
2VNU
 
 | | Crystal structure of Sc Rrp44 | | Descriptor: | 5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP)-3', EXOSOME COMPLEX EXONUCLEASE RRP44, MAGNESIUM ION, ... | | Authors: | Lorentzen, E, Basquin, J, Conti, E. | | Deposit date: | 2008-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active Subunit of the Yeast Exosome Core, Rrp44: Diverse Modes of Substrate Recruitment in the Rnase II Nuclease Family
Mol.Cell, 29, 2008
|
|
4W8V
 
 | |
4W8X
 
 | | Crystal Structure of Cmr1 from Pyrococcus furiosus bound to a nucleotide | | Descriptor: | CRISPR system Cmr subunit Cmr1-1, GUANOSINE-3'-MONOPHOSPHATE, PHOSPHATE ION | | Authors: | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
4W8Y
 
 | | Structure of full length Cmr2 from Pyrococcus furiosus (Manganese bound form) | | Descriptor: | CRISPR system Cmr subunit Cmr2, MANGANESE (II) ION, ZINC ION | | Authors: | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
4WFD
 
 | | Structure of the Rrp6-Rrp47-Mtr4 interaction | | Descriptor: | ATP-dependent RNA helicase DOB1, Exosome complex exonuclease RRP6, Exosome complex protein LRP1, ... | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4WFC
 
 | | Structure of the Rrp6-Rrp47 interaction | | Descriptor: | Exosome complex exonuclease RRP6, Exosome complex protein LRP1, SULFATE ION | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4XR7
 
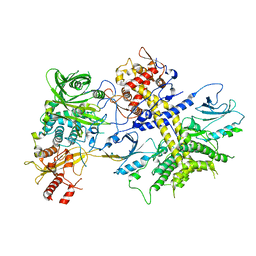 | | Structure of the Saccharomyces cerevisiae PAN2-PAN3 core complex | | Descriptor: | PAB-dependent poly(A)-specific ribonuclease subunit PAN2, PAB-dependent poly(A)-specific ribonuclease subunit PAN3 | | Authors: | Schafer, I.B, Rode, M, Bonneau, F, Schussler, S, Conti, E. | | Deposit date: | 2015-01-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.796 Å) | | Cite: | The structure of the Pan2-Pan3 core complex reveals cross-talk between deadenylase and pseudokinase.
Nat. Struct. Mol. Biol., 21, 2014
|
|
4N0L
 
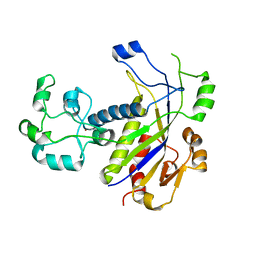 | | Methanopyrus kandleri Csm3 crystal structure | | Descriptor: | Predicted component of a thermophile-specific DNA repair system, contains a RAMP domain, ZINC ION | | Authors: | Hrle, A, Su, A.A, Ebert, J, Benda, C, Conti, E, Randau, L. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure and RNA-binding properties of the Type III-A CRISPR-associated protein Csm3.
Rna Biol., 10, 2013
|
|
6H25
 
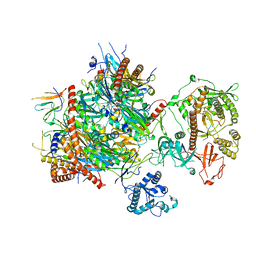 | | Human nuclear RNA exosome EXO-10-MPP6 complex | | Descriptor: | Exosome complex component CSL4, Exosome complex component MTR3, Exosome complex component RRP4, ... | | Authors: | Gerlach, P, Schuller, J.M, Falk, S, Basquin, J, Conti, E. | | Deposit date: | 2018-07-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Distinct and evolutionary conserved structural features of the human nuclear exosome complex.
Elife, 7, 2018
|
|
2BR2
 
 | | RNase PH core of the archaeal exosome | | Descriptor: | CHLORIDE ION, EXOSOME COMPLEX EXONUCLEASE 1, EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | lorentzen, E, Fribourg, S, Conti, E. | | Deposit date: | 2005-04-30 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Archaeal Exosome Core is a Hexameric Ring Structure with Three Catalytic Subunits.
Nat.Struct.Mol.Biol., 12, 2005
|
|
4Q8H
 
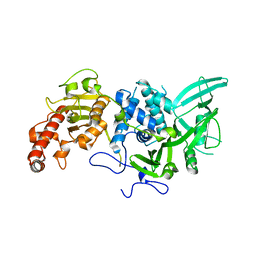 | |
4Q8G
 
 | |
7Z4Z
 
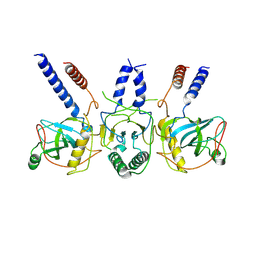 | | Human NEXT dimer - focused reconstruction of the dimerization module | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7Z4Y
 
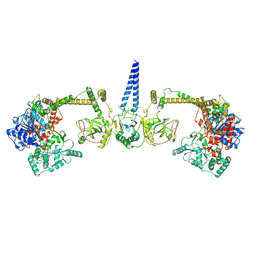 | | Human NEXT dimer - overall reconstruction of the core complex | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
