1YFZ
 
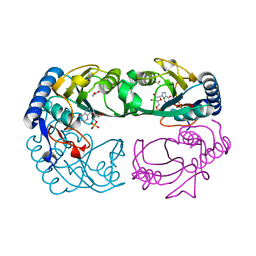 | | Novel IMP Binding in Feedback Inhibition of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis | | Descriptor: | ACETATE ION, Hypoxanthine-guanine phosphoribosyltransferase, INOSINIC ACID, ... | | Authors: | Chen, Q, Liang, Y, Su, X, Gu, X, Zheng, X, Luo, M. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative IMP Binding in Feedback Inhibition of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis.
J.Mol.Biol., 348, 2005
|
|
4Z90
 
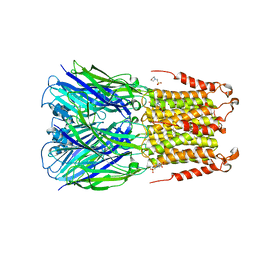 | | ELIC bound with the anesthetic isoflurane in the resting state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Chen, Q, Kinde, M, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
4Z91
 
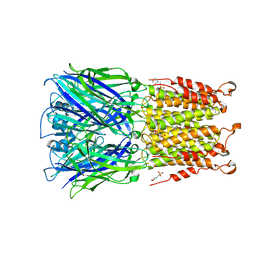 | | ELIC cocrystallized with isofluorane in a desensitized state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 1.7.6 3-bromanylpropan-1-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Chen, Q, Kinde, M.N, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3915 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
5ZB7
 
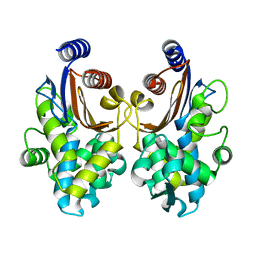 | | CTX-M-64 apoenzyme | | Descriptor: | Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-02-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
7MTB
 
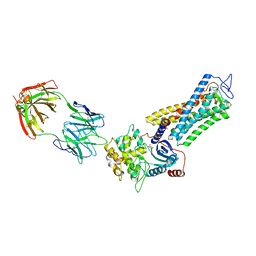 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab6 | | Descriptor: | Fab6 heavy chain, Fab6 light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT8
 
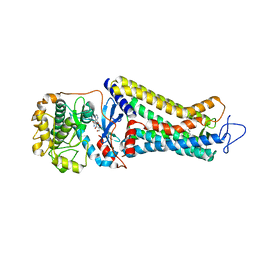 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MTA
 
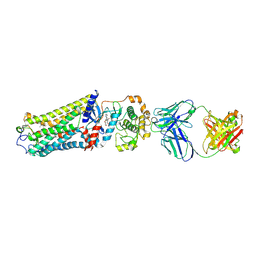 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab1 | | Descriptor: | Fab1 Heavy chain, Fab1 Light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT9
 
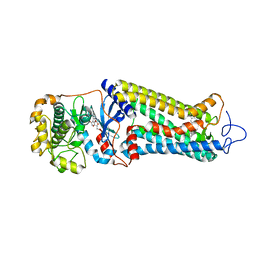 | | Rhodopsin kinase (GRK1) in complex with rhodopsin | | Descriptor: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
5SXU
 
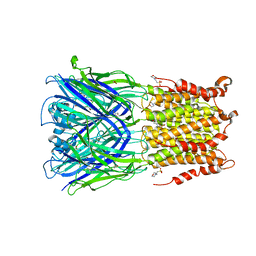 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a desensitized state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, 3-AMINOPROPANE, ... | | Authors: | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
5SXV
 
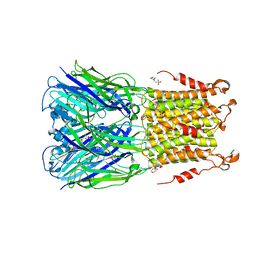 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a resting state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, Cys-loop ligand-gated ion channel | | Authors: | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
5TV1
 
 | | active arrestin-3 with inositol hexakisphosphate | | Descriptor: | Beta-arrestin-2, GLYCEROL, INOSITOL HEXAKISPHOSPHATE | | Authors: | Chen, Q, Gilbert, N.C, Perry, N.A, Vishniveteskiy, S, Gurevich, V.V, Iverson, T.M. | | Deposit date: | 2016-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of arrestin-3 activation and signaling.
Nat Commun, 8, 2017
|
|
6CPZ
 
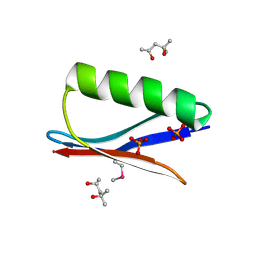 | | Selenomethionine mutant (I6Sem) of protein GB1 examined by X-ray diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-14 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6CHE
 
 | |
6CTE
 
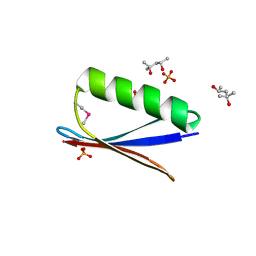 | | 77Se-NMR probes the protein environment of selenomethionine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-22 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6C9O
 
 | |
6D1S
 
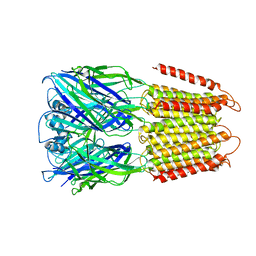 | | Crystal structure of an apo chimeric human alpha1GABAA receptor | | Descriptor: | chimeric alpha1GABAA receptor | | Authors: | Chen, Q, Arjunan, P, Cohen, A.E, Xu, Y, Tang, P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of neurosteroid anesthetic action on GABAAreceptors.
Nat Commun, 9, 2018
|
|
6CNE
 
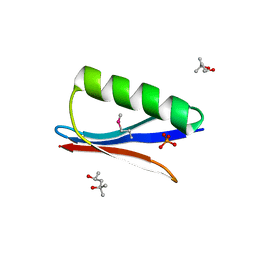 | | Selenomethionine variant (V29SeM) of protein GB1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, PHOSPHATE ION | | Authors: | Chen, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6CDU
 
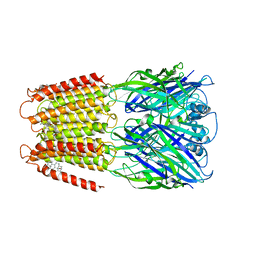 | | Crystal structure of a chimeric human alpha1GABAA receptor in complex with alphaxalone | | Descriptor: | (3a,5a)-3-Hydroxypregnane-11,20-dione, chimeric alpha1GABAA receptor | | Authors: | Chen, Q, Arjunan, P, Cohen, A.E, Xu, Y, Tang, P. | | Deposit date: | 2018-02-09 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural basis of neurosteroid anesthetic action on GABAAreceptors.
Nat Commun, 9, 2018
|
|
6LDU
 
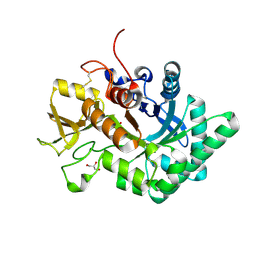 | |
6LE7
 
 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2 | | Descriptor: | (4R)-3-(2-hydroxyphenyl)-4-(3-methoxy-4-propoxy-phenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, Probable endochitinase | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85753441 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2
To Be Published
|
|
6LE8
 
 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 1 | | Descriptor: | (4R)-4-(4-ethoxyphenyl)-3-(2-hydroxyphenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39909327 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 1
To Be Published
|
|
5WPT
 
 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed II conformation at 3.75 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
5WPQ
 
 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed I conformation at 3.64 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
5WPV
 
 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs at 3.59 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
3LCY
 
 | |
