7KO4
 
 | |
7KOR
 
 | |
7KO7
 
 | |
7KON
 
 | |
7KQ7
 
 | | Crystal structure of IL21R in complex with an antibody Fab fragment | | Descriptor: | Antibody heavy chain, Antibody light chain, Interleukin-21 receptor | | Authors: | Mosyak, L, Svenson, K. | | Deposit date: | 2020-11-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Combining random mutagenesis, structure-guided design and next-generation sequencing to mitigate polyreactivity of an anti-IL-21R antibody.
Mabs, 13, 2021
|
|
1BH4
 
 | |
8SXS
 
 | |
7L6O
 
 | | Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 - gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2020-12-23 | | Release date: | 2021-04-14 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
1FKK
 
 | | ATOMIC STRUCTURE OF FKBP12, AN IMMUNOPHILIN BINDING PROTEIN | | Descriptor: | FK506 BINDING PROTEIN, SULFATE ION | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1BFB
 
 | | BASIC FIBROBLAST GROWTH FACTOR COMPLEXED WITH HEPARIN TETRAMER FRAGMENT | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Faham, S, Rees, D.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Heparin structure and interactions with basic fibroblast growth factor.
Science, 271, 1996
|
|
7JUT
 
 | |
8SMI
 
 | | Crystal structure of antibody WRAIR-2123 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2123 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SGU
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-13 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
1BC1
 
 | | RECOMBINANT RAT ANNEXIN V, QUADRUPLE MUTANT (T72K, S144K, S228K, S303K) | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
7JUV
 
 | | Crystal Structure of KSR2:MEK1 in complex with AMP-PNP, and allosteric MEK inhibitor APS-9-95-1 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 2, MAGNESIUM ION, ... | | Authors: | Khan, Z.M, Dar, A.C, Scopton, A.P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural basis for the action of the drug trametinib at KSR-bound MEK.
Nature, 588, 2020
|
|
1BCZ
 
 | | RECOMBINANT RAT ANNEXIN V, T72S MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BCW
 
 | | RECOMBINANT RAT ANNEXIN V, T72A MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BC3
 
 | | RECOMBINANT RAT ANNEXIN V, TRIPLE MUTANT (T72K, S144K, S228K) | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
8SMW
 
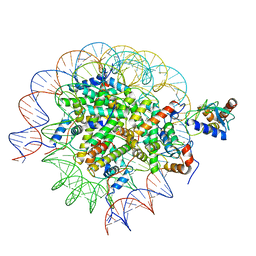 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN7
 
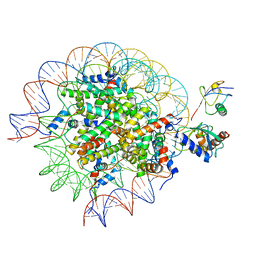 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 5) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SMX
 
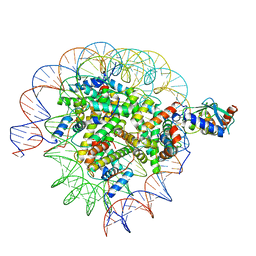 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN6
 
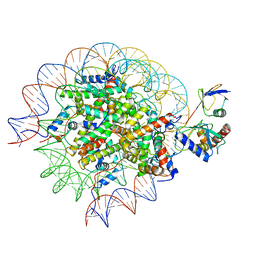 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 4) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8T5C
 
 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
8SN2
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c (UbcH5c chemically conjugated to histone H2A) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN5
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 3) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
