8IJK
 
 | | human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
6GHZ
 
 | |
6GI3
 
 | |
6GHY
 
 | |
4W2R
 
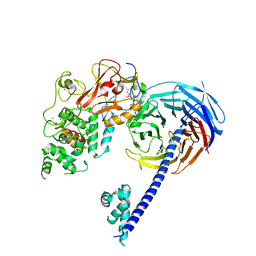 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
7ME4
 
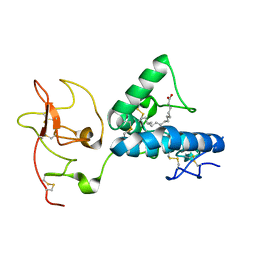 | | Structure of the extracellular WNT-binding module in Drosophila Ror2/Nrk | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOLEIC ACID, Tyrosine-protein kinase transmembrane receptor Ror2 | | Authors: | Mendrola, J.M, Shi, F, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
6IFE
 
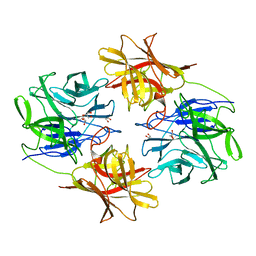 | | A Glycoside Hydrolase Family 43 beta-Xylosidase | | Descriptor: | Beta-xylosidase, GLYCEROL | | Authors: | Li, N, Liu, Y, Zhang, R, Zhou, J.P, Huang, Z.X. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 beta-xylosidase: Activity and instability at high neutral salt concentrations.
Food Chem, 301, 2019
|
|
7ME5
 
 | | Structure of the extracellular WNT-binding module in Drl-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tyrosine-protein kinase transmembrane receptor DRL-2 | | Authors: | Shi, F, Mendrola, J.M, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
7MXN
 
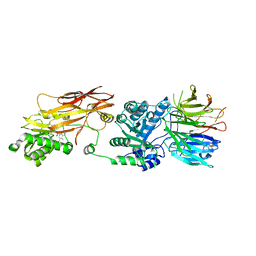 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06939999 | | Descriptor: | (1S,2S,3S,5R)-3-{[6-(difluoromethyl)-5-fluoro-1,2,3,4-tetrahydroisoquinolin-8-yl]oxy}-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MX7
 
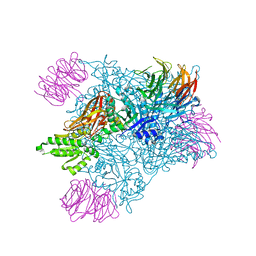 | | PRMT5:MEP50 complexed with inhibitor PF-06939999 | | Descriptor: | (1S,2S,3S,5R)-3-{[6-(difluoromethyl)-5-fluoro-1,2,3,4-tetrahydroisoquinolin-8-yl]oxy}-5-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MXA
 
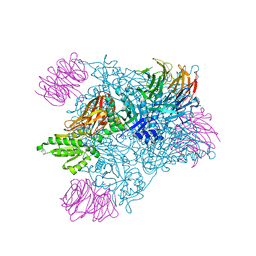 | | PRMT5:MEP50 complexed with inhibitor PF-06855800 | | Descriptor: | 7-[(5R)-5-C-(4-chloro-3-fluorophenyl)-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MXC
 
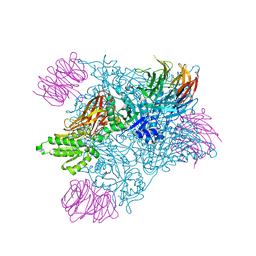 | | PRMT5:MEP50 complexed with adenosine | | Descriptor: | ADENOSINE, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7YZY
 
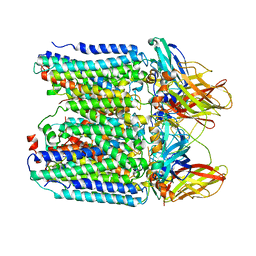 | | pMMO structure from native membranes by cryoET and STA | | Descriptor: | Methane monooxygenase subunit C2, Particulate methane monooxygenase alpha subunit, Particulate methane monooxygenase beta subunit | | Authors: | Zhu, Y, Ni, T, Zhang, P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-10 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and activity of particulate methane monooxygenase arrays in methanotrophs.
Nat Commun, 13, 2022
|
|
6M08
 
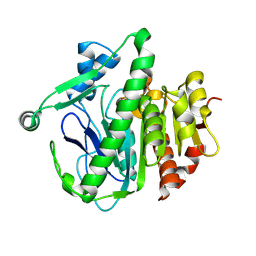 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor. | | Descriptor: | (2S)-2-[(Z)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
6M32
 
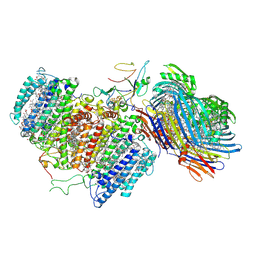 | | Cryo-EM structure of FMO-RC complex from green sulfur bacteria | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Chen, J.H, Zhang, X. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the photosynthetic complex from a green sulfur bacterium.
Science, 370, 2020
|
|
8HJV
 
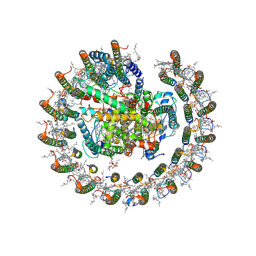 | | Cryo-EM structure of carotenoid-depleted RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8HJU
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
6M07
 
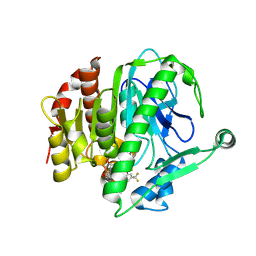 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(E)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)-2-[2,2,2-tris(fluoranyl)ethoxy]phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
6M06
 
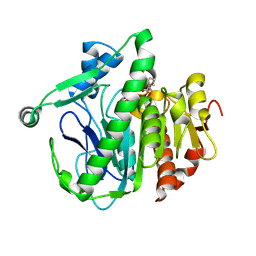 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(Z)-1,3-bis(oxidanyl)-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
4XDP
 
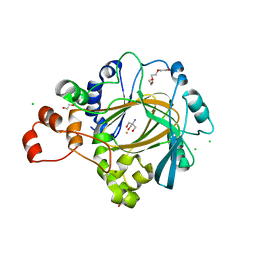 | | Crystal structure of human KDM4C catalytic domain bound to tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A High-Throughput Mass Spectrometry Assay Coupled with Redox Activity Testing Reduces Artifacts and False Positives in Lysine Demethylase Screening.
J Biomol Screen, 20, 2015
|
|
6ML6
 
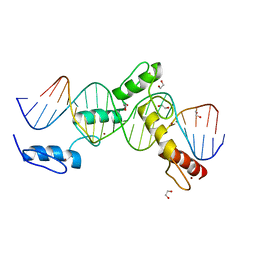 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpA 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*(5CM)P*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
4XDO
 
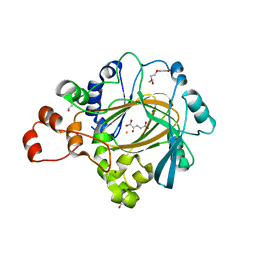 | | Crystal structure of human KDM4C catalytic domain with OGA | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Lysine-specific demethylase 4C, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A High-Throughput Mass Spectrometry Assay Coupled with Redox Activity Testing Reduces Artifacts and False Positives in Lysine Demethylase Screening.
J Biomol Screen, 20, 2015
|
|
6ML5
 
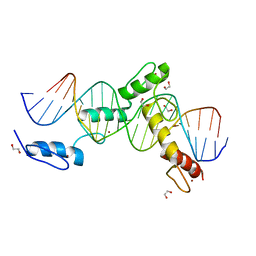 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML4
 
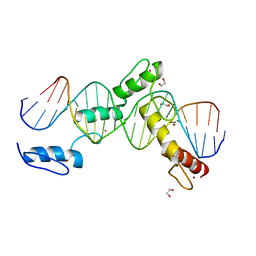 | | BTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 3) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML7
 
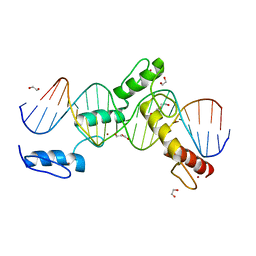 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpG 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*(5CM)P*GP*AP*AP*TP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
