3N79
 
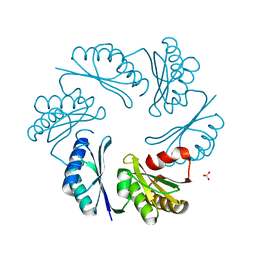 | | PduT C38S Mutant from Salmonella enterica Typhimurium | | Descriptor: | CHLORIDE ION, PduT, SODIUM ION, ... | | Authors: | Crowley, C.S, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insight into the Mechanisms of Transport across the Salmonella enterica Pdu Microcompartment Shell.
J.Biol.Chem., 285, 2010
|
|
3NH4
 
 | | Crystal structure of murine aminoacylase 3 | | Descriptor: | ACETATE ION, Aspartoacylase-2, CESIUM ION, ... | | Authors: | Hsieh, J.M, Tsirulnikov, K, Sawaya, M.R, Magilnick, N, Abuladze, N, Kurtz, I, Abramson, J, Pushkin, A. | | Deposit date: | 2010-06-14 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of aminoacylase 3 in complex with acetylated substrates.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MLG
 
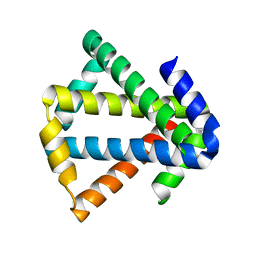 | | 2ouf-2x, a designed knotted protein | | Descriptor: | 2X chimera of Helicobacter pylori protein HP0242 | | Authors: | King, N.P, Sawaya, M.R, Jacobitz, A.W, Yeates, T.O. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure and folding of a designed knotted protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NH8
 
 | | Crystal structure of murine aminoacylase 3 in complex with N-acetyl-S-1,2-dichlorovinyl-L-cysteine | | Descriptor: | Aspartoacylase-2, CHLORIDE ION, N-acetyl-S-[(1S)-1,2-dichloroethyl]-L-cysteine, ... | | Authors: | Hsieh, J.M, Tsirulnikov, K, Sawaya, M.R, Magilnick, N, Abuladze, N, Kurtz, I, Abramson, J, Pushkin, A. | | Deposit date: | 2010-06-14 | | Release date: | 2010-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structures of aminoacylase 3 in complex with acetylated substrates.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NH5
 
 | | Crystal structure of E177A-mutant murine aminoacylase 3 | | Descriptor: | ACETATE ION, Aspartoacylase-2, CHLORIDE ION, ... | | Authors: | Hsieh, J.M, Tsirulnikov, K, Sawaya, M.R, Magilnick, N, Abuladze, N, Kurtz, I, Abramson, J, Pushkin, A. | | Deposit date: | 2010-06-14 | | Release date: | 2010-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structures of aminoacylase 3 in complex with acetylated substrates.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NFZ
 
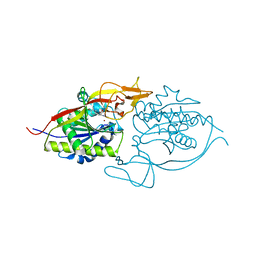 | | Crystal structure of murine aminoacylase 3 in complex with N-acetyl-L-tyrosine | | Descriptor: | Aspartoacylase-2, CHLORIDE ION, N-acetyl-L-tyrosine, ... | | Authors: | Hsieh, J.M, Tsirulnikov, K, Sawaya, M.R, Magilnick, N, Abuladze, N, Kurtz, I, Abramson, J, Pushkin, A. | | Deposit date: | 2010-06-10 | | Release date: | 2010-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structures of aminoacylase 3 in complex with acetylated substrates.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NGG
 
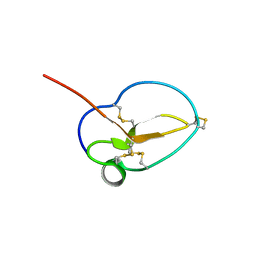 | | X-ray Structure of Omwaprin | | Descriptor: | Omwaprin-a | | Authors: | Banigan, J.R, Mandal, K, Sawaya, M.R, Thammavongsa, V, Hendrickx, A.P.A, Schneewind, O, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography.
Protein Sci., 19, 2010
|
|
4R0U
 
 | |
4RBT
 
 | |
4R0P
 
 | |
4RBV
 
 | |
4RBU
 
 | |
4QXX
 
 | |
4RC1
 
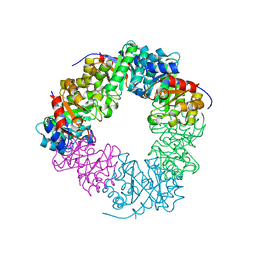 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii with PRPP | | Descriptor: | PHOSPHATE ION, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E.J, Cascio, D, Sawaya, M.R, Yeates, T.O, Rasche, M.E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4R0W
 
 | |
4RIK
 
 | | Amyloid forming segment, AVVTGVTAV, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 69-77 | | Descriptor: | Alpha-synuclein | | Authors: | Guenther, E.L, Sawaya, M.R, Ivanova, M, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
4RIL
 
 | | Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.43 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
4RKP
 
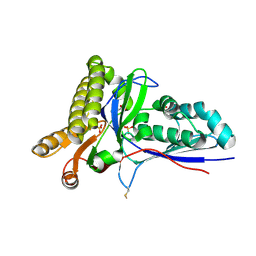 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (apo form) | | Descriptor: | ACETATE ION, Putative uncharacterized protein Ta1305, SULFATE ION | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RKZ
 
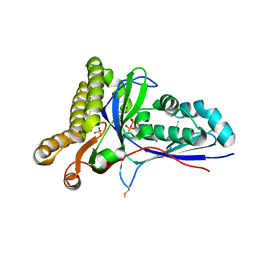 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (Mevalonate 3-Phosphate/ADP Bound) | | Descriptor: | (3R)-5-hydroxy-3-methyl-3-(phosphonooxy)pentanoic acid, ADENOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein Ta1305, ... | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RKS
 
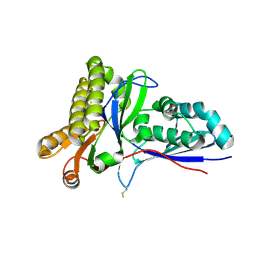 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (Mevalonate Bound) | | Descriptor: | (R)-MEVALONATE, ACETATE ION, GLYCEROL, ... | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | Descriptor: | GLYCEROL, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-27 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4RPM
 
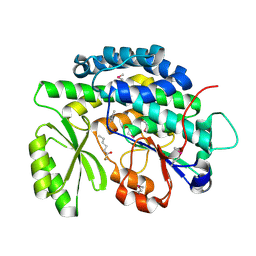 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM with bound hexanoyl | | Descriptor: | HEXANOIC ACID, HEXANOYL-COENZYME A, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-30 | | Release date: | 2015-09-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4UNU
 
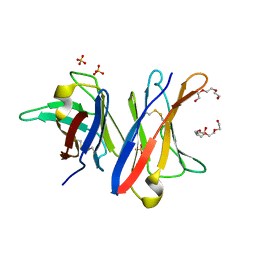 | | MCG - a dimer of lambda variable domains | | Descriptor: | IG LAMBDA CHAIN V-II REGION MGC, POLYETHYLENE GLYCOL (N=34), SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Landau, M, Ryan, C, Whitelegge, J, Casio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Formation of Amyloid Fibers by Monomeric Light-Chain Variable Domains.
J.Biol.Chem., 289, 2014
|
|
4UNT
 
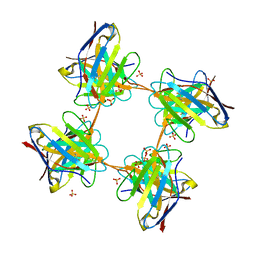 | | Induced monomer of the Mcg variable domain | | Descriptor: | IG LAMBDA CHAIN V-II REGION MGC, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Landau, M, Ryan, C.M, Whitelegge, J.P, Phillips, M.L, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Formation of Amyloid Fibers by Monomeric Light-Chain Variable Domains.
J.Biol.Chem., 289, 2014
|
|
4U9P
 
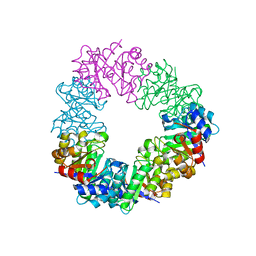 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii | | Descriptor: | GLYCEROL, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E, Shin, A, Cascio, D, Sawaya, M.R, Arbing, M, Rasche, M.E. | | Deposit date: | 2014-08-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
