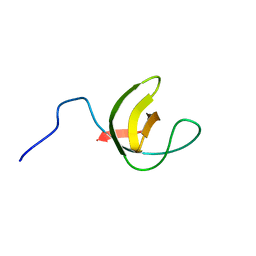
3LPR
 
 | |
8FM6
 
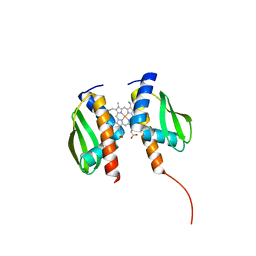 | |
1LGB
 
 | |
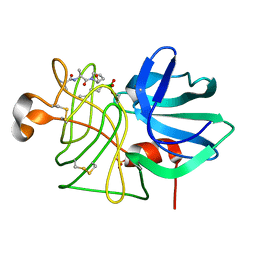
6T66
 
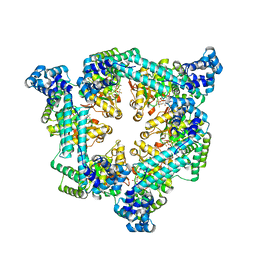 | | Crystal structure of the Vibrio cholerae replicative helicase (DnaB) with GDP-AlF4 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Replicative DNA helicase, ... | | Authors: | Legrand, P, Quevillon-Cheruel, S, Li de la Sierra-Gallay, I, Walbott, H. | | Deposit date: | 2019-10-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
Nucleic Acids Res., 49, 2021
|
|
2MIO
 
 | | Solution NMR Structure of SH3 Domain 1 of Rho GTPase-activating Protein 10 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9129A | | Descriptor: | Rho GTPase-activating protein 10 | | Authors: | Xu, X, Eletsky, A, Pulavarti, S.V.S.R.K, Wang, H, Janjua, H, Xiao, R, Everett, J.K, Sukumaran, D.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of SH3 Domain 1 of Rho GTPase-activating Protein 10 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9129A
To be Published
|
|
7PSZ
 
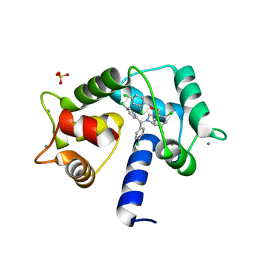 | | Crystal structure of CaM in complex with CDZ (form 1) | | Descriptor: | 1-[bis(4-chlorophenyl)methyl]-3-[(2~{R})-2-(2,4-dichlorophenyl)-2-[(2,4-dichlorophenyl)methoxy]ethyl]imidazole, CALCIUM ION, Calmodulin-1, ... | | Authors: | Mechaly, A.E, Leger, C, Haouz, A, Chenal, A. | | Deposit date: | 2021-09-24 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
Bmc Biol., 20, 2022
|
|
7PU9
 
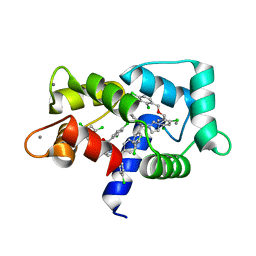 | | Crystal structure of CaM in complex with CDZ (form 2) | | Descriptor: | 1-[bis(4-chlorophenyl)methyl]-3-[(2~{R})-2-(2,4-dichlorophenyl)-2-[(2,4-dichlorophenyl)methoxy]ethyl]imidazole, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Leger, C, Haouz, A, Chenal, A. | | Deposit date: | 2021-09-28 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
Bmc Biol., 20, 2022
|
|
4XAH
 
 | |
7QK0
 
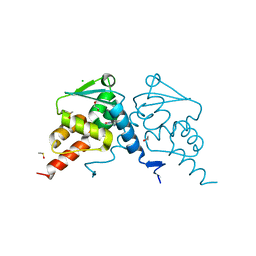 | | Crystal structure of human BCL6 BTB domain in complex with compound 12a | | Descriptor: | (2~{S})-10-[[5-chloranyl-2-[(3~{S},5~{R})-3-methyl-5-oxidanyl-piperidin-1-yl]pyrimidin-4-yl]amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Gunnell, E.A, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Improved Binding Affinity and Pharmacokinetics Enable Sustained Degradation of BCL6 In Vivo .
J.Med.Chem., 65, 2022
|
|
6S84
 
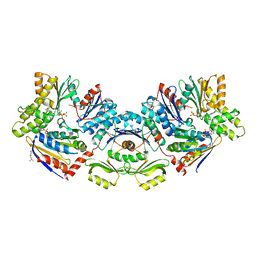 | | TsaBDE complex from Thermotoga maritima | | Descriptor: | ATPase YjeE, predicted to have essential role in cell wall biosynthesis, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Missoury, S, Li-de-La-Sierra-Gallay, I, van Tilbeurgh, H. | | Deposit date: | 2019-07-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structure of the TsaB/TsaD/TsaE complex reveals an unexpected mechanism for the bacterial t6A tRNA-modification.
Nucleic Acids Res., 46, 2018
|
|
2IVY
 
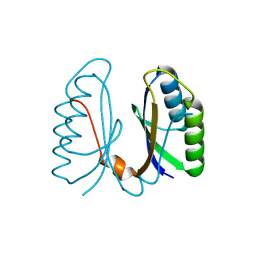 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
3LTM
 
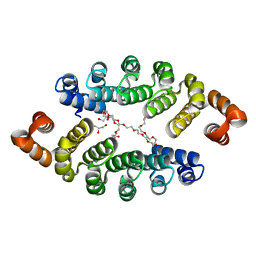 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | Alpha-Rep4, DODECAETHYLENE GLYCOL, GLYCEROL, ... | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
3LTJ
 
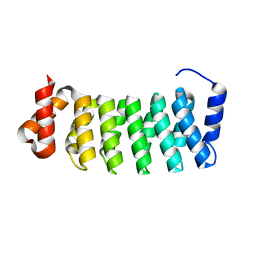 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | AlphaRep-4 | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
2JRB
 
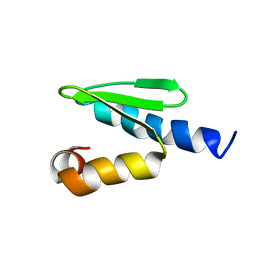 | | C-terminal domain of ORF1p from mouse LINE-1 | | Descriptor: | ORF 1 protein | | Authors: | Januszyk, K, Clubb, R. | | Deposit date: | 2007-06-21 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1.
J.Biol.Chem., 282, 2007
|
|
4TRF
 
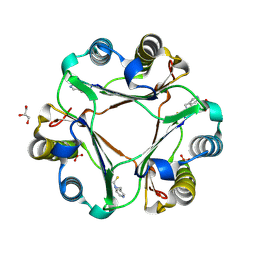 | |
3PQU
 
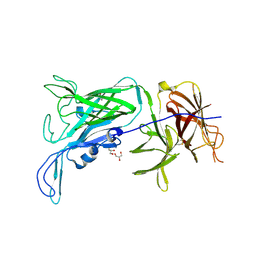 | |
4ZV6
 
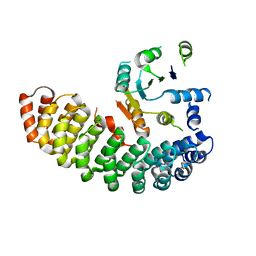 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | Descriptor: | AlphaRep-7, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
5O7N
 
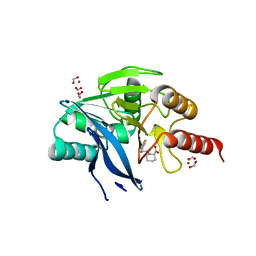 | | Beta-lactamase VIM-2 in complex with (2R)-1-(2-Benzyl-3-mercaptopropanoyl)piperidine-2-carboxylic acid | | Descriptor: | (2~{R})-1-[(2~{S})-2-(phenylmethyl)-3-sulfanyl-propanoyl]piperidine-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase VIM-2, ... | | Authors: | Buettner, D, Kramer, J.S, Pogoryelov, D, Proschak, E. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Challenges in the Development of a Thiol-Based Broad-Spectrum Inhibitor for Metallo-beta-Lactamases.
Acs Infect Dis., 4, 2018
|
|
3C6B
 
 | |
6T9Q
 
 | |
4TRU
 
 | |
2XNT
 
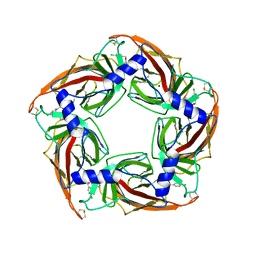 | | Acetylcholine binding protein (AChBP) as template for hierarchical in silico screening procedures to identify structurally novel ligands for the nicotinic receptors | | Descriptor: | (2S)-2-[(4-CHLOROBENZYL)OXY]-2-PHENYLETHANAMINE, BROMIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Rucktooa, P, Akdemir, A, deEsch, I, Sixma, T.K. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Acetylcholine Binding Protein (Achbp) as Template for Hierarchical in Silico Screening Procedures to Identify Structurally Novel Ligands for the Nicotinic Receptors.
Bioorg.Med.Chem., 19, 2011
|
|
3UQS
 
 | |
4RUD
 
 | | Crystal structure of a three finger toxin | | Descriptor: | Three-finger toxin 3b, ZINC ION | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2014-11-18 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fulditoxin, representing a new class of dimeric snake toxins, defines novel pharmacology at nicotinic ACh receptors.
Br.J.Pharmacol., 177, 2020
|
|
2JG5
 
 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
