7OPB
 
 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
6ZOQ
 
 | | Oestrogen receptor ligand binding domain in complex with compound 16 | | Descriptor: | Estrogen receptor, ~{N}-[4-[(6~{S},8~{R})-7-[(1-fluoranylcyclopropyl)methyl]-8-methyl-2,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-3-methoxy-phenyl]-1-(3-fluoranylpropyl)azetidin-3-amine | | Authors: | Breed, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
6ZOR
 
 | | Oestrogen receptor ligand binding domain in complex with compound 28 | | Descriptor: | 6-[(6~{S},8~{R})-8-methyl-7-[2,2,2-tris(fluoranyl)ethyl]-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-~{N}-(1-propylazetidin-3-yl)pyridin-3-amine, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
6ZOS
 
 | | Oestrogen receptor ligand binding domain in complex with compound 18 | | Descriptor: | 6-[(6~{S},8~{R})-7-[(1-fluoranylcyclopropyl)methyl]-8-methyl-2,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-~{N}-[1-(3-fluoranylpropyl)azetidin-3-yl]pyridin-3-amine, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
6C0S
 
 | |
3JZN
 
 | | Structure of EED in apo form | | Descriptor: | Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Ouyang, H, Qiu, W, MacKenzie, F, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5Q0E
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(4S,8S)-8-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-4-methyl-2-oxo-1,3,4,5,6,7,8,10-octahydro-2H-12,9-(azeno)-1,10-benzodiazacyclotetradecin-15-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-12 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Macrocyclic inhibitors of Factor XIa: Discovery of alkyl-substituted macrocyclic amide linkers with improved potency.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3FC3
 
 | | Crystal structure of the beta-beta-alpha-Me type II restriction endonuclease Hpy99I | | Descriptor: | 5'-(*DCP*DTP*DCP*DGP*DAP*DCP*DGP*DTP*DAP*DGP*DA)-3', 5'-(*DTP*DAP*DCP*DGP*DTP*DCP*DGP*DAP*DGP*DTP*DC)-3', Restriction endonuclease Hpy99I, ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Nucleic Acids Res., 37, 2009
|
|
7S5B
 
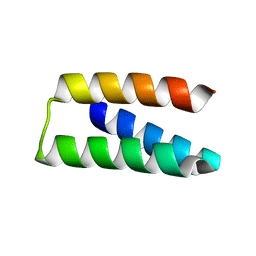 | |
7RDH
 
 | |
6INI
 
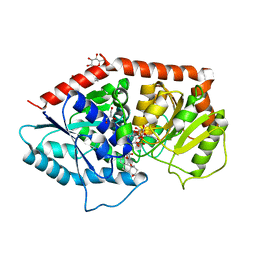 | | a glycosyltransferase complex with UDP and the product | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
7TO4
 
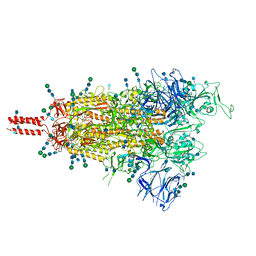 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2022-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
7TNW
 
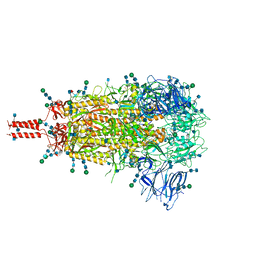 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2022-01-21 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
3K26
 
 | | Complex structure of EED and trimethylated H3K4 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Bian, C.B, Xu, C, Qiu, W, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3K27
 
 | | Complex structure of EED and trimethylated H3K9 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Bian, C.B, Xu, C, Qiu, W, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6INF
 
 | | a glycosyltransferase complex with UDP | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
7QVL
 
 | | OESTROGEN RECEPTOR LIGAND BINDING DOMAIN IN COMPLEX WITH COMPOUND 38 | | Descriptor: | (2~{R})-3-[(1~{R},3~{R})-1-[5-fluoranyl-2-[2-(3-fluoranylpropylamino)ethoxy]-3-methyl-pyridin-4-yl]-3-methyl-1,3,4,9-tetrahydropyrido[3,4-b]indol-2-yl]-2-methyl-propanoic acid, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Zwitterionic Series of Selective Estrogen Receptor Degrader-Antagonists.
J.Med.Chem., 66, 2023
|
|
7QVJ
 
 | | ESTROGEN RECEPTOR ALPHA IN COMPLEX WITH COMPOUND 29 | | Descriptor: | 2,2-bis(fluoranyl)-3-[(1~{R},3~{R})-1-[6-fluoranyl-3-[2-(3-fluoranylpropylamino)ethoxy]-2-methyl-phenyl]-3-methyl-1,3,4,9-tetrahydropyrido[3,4-b]indol-2-yl]propan-1-ol, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Zwitterionic Series of Selective Estrogen Receptor Degrader-Antagonists.
J.Med.Chem., 66, 2023
|
|
5NCY
 
 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-(3-methyl-5-oxidanylidene-6-phenyl-[1,3]thiazolo[3,2-a]pyridin-7-yl)ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ... | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
5NCZ
 
 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-[6-[3-[(dimethylamino)methyl]phenyl]-3-methyl-5-oxidanylidene-[1,3]thiazolo[3,2-a]pyridin-7-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
7MBO
 
 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.924 Å) | | Cite: | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
3JZH
 
 | | EED-H3K79me3 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JPX
 
 | | EED: A Novel Histone Trimethyllysine Binder Within The EED-EZH2 Polycomb Complex | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JZG
 
 | | Structure of EED in complex with H3K27me3 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6INH
 
 | | A glycosyltransferase with UDP and the substrate | | Descriptor: | 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, UDP-glycosyltransferase 76G1, ... | | Authors: | Zhu, X. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
