3PRE
 
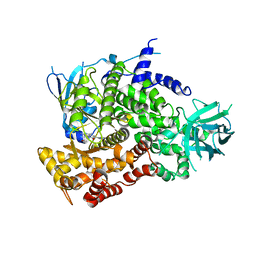 | | Quinazolines with intra-molecular hydrogen bonding scaffold (iMHBS) as PI3K/mTOR dual inhibitors. | | Descriptor: | 2-amino-8-(trans-4-methoxycyclohexyl)-4-methyl-6-(1H-pyrazol-3-yl)pyrido[2,3-d]pyrimidin-7(8H)-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Knighton, D.R, Greasley, S.E, Rodgers, C.M.-L. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Quinazolines with intra-molecular hydrogen bonding scaffold (iMHBS) as PI3K/mTOR dual inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
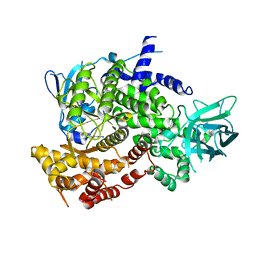
3PRZ
 
 | |
5URA
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP7 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, brain, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85002172 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
5UR9
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP5 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, epidermal, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19800353 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
7WV9
 
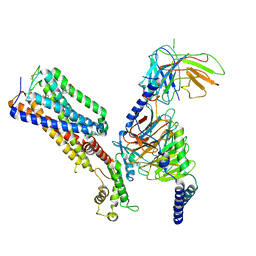 | | Allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 in complex with Gi protein | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, ... | | Authors: | Xu, Z, Shao, Z. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
1M3G
 
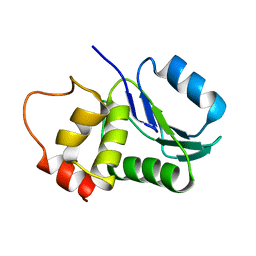 | |
7VGC
 
 | |
5U6Z
 
 | |
7VGB
 
 | | Crystal structure of apo prolyl oligopeptidase from Microbulbifer arenaceous | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Huang, P, Jiang, Z.Q. | | Deposit date: | 2021-09-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | The structure and molecular dynamics of prolyl oligopeptidase from Microbulbifer arenaceous provide insights into catalytic and regulatory mechanisms.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4UVE
 
 | | Discovery of pyrimidine isoxazoles InhA in complex with compound 9 | | Descriptor: | 2-(4,6-DIMETHYLPYRIMIDIN-2-YL)SULFANYLETHANOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], MAGNESIUM ION, ... | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Ghorpade, S, Cowan, S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Hitting the Target in More Than One Way: Novel, Direct Inhibitors of Mycobacterium Tuberculosis Enoyl Acp Reductase
To be Published
|
|
4UVG
 
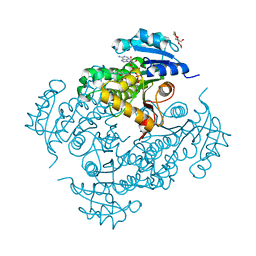 | | Discovery of pyrimidine isoxazoles InhA in complex with compound 15 | | Descriptor: | 5-[(4,6-dimethylpyrimidin-2-yl)sulfanylmethyl]isoxazole-3-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], MAGNESIUM ION, ... | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Ghorpade, S, Cowan, S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hitting the Target in More Than One Way: Novel, Direct Inhibitors of Mycobacterium Tuberculosis Enoyl Acp Reductase
To be Published
|
|
5KVT
 
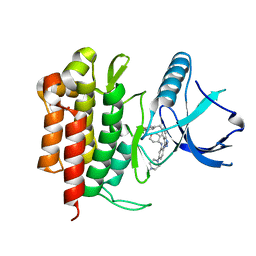 | | THE STRUCTURE OF TRKA KINASE DOMAIN BOUND TO THE INHIBITOR ENTRECTINIB | | Descriptor: | Entrectinib, GLYCEROL, High affinity nerve growth factor receptor | | Authors: | Jin, L, Yan, S, Wei, G, Li, G, Harris, J, Vernier, J.-M. | | Deposit date: | 2016-07-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antitumor Activity and Safety of the Pan-TRK, ROS1, and ALK inhibitor Entrectinib (RXDX-101): Combined Results from Two Phase I Trials
To Be Published
|
|
5H5P
 
 | |
4ZZ3
 
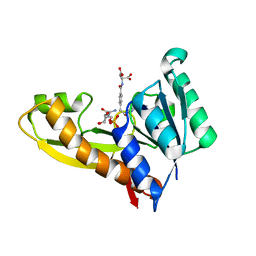 | | Human GAR transformylase in complex with GAR and pemetrexed | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[2-(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)ethyl]benzoyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
4ZZ1
 
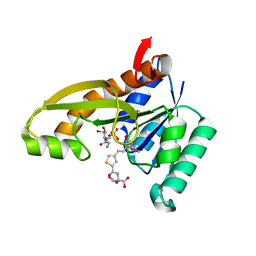 | | HUMAN GAR TRANSFORMYLASE IN COMPLEX WITH GAR AND (S)-2-({4-[3-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-6-YL)-PROPYL]-THIOPHENE-2-CARBONYL}-AMINO)-PENTANEDIOIC ACID | | Descriptor: | (S)-2-({4-[3-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)-propyl]-thiophene-2-carbonyl}-amino)-pentanedioic acid, GLYCINAMIDE RIBONUCLEOTIDE, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
4ZZ2
 
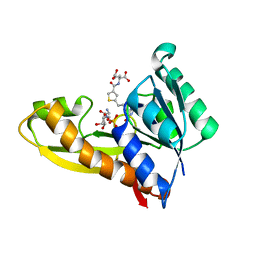 | | HUMAN GAR TRANSFORMYLASE IN COMPLEX WITH GAR AND (S)-2-({5-[3-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-6-YL)-PROPYL]-THIOPHENE-3-CARBONYL}-AMINO)-PENTANEDIOIC ACID | | Descriptor: | (S)-2-({5-[3-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)-propyl]-thiophene-3-carbonyl}-amino)-pentanedioic acid, GLYCINAMIDE RIBONUCLEOTIDE, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
2AF5
 
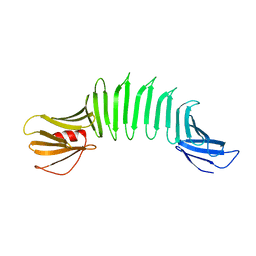 | | 2.5A X-ray Structure of Engineered OspA protein | | Descriptor: | Engineered Outer Surface Protein A (OspA) with the inserted two beta-hairpins | | Authors: | Makabe, K, Mcelheny, D, Tereshko, V, Hilyard, A, Koide, A, Koide, S. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structures of peptide self-assembly mimics.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HKD
 
 | |
2I5V
 
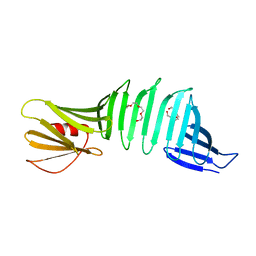 | | Crystal structure of OspA mutant | | Descriptor: | Outer surface protein A, TETRAETHYLENE GLYCOL | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2006-08-25 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Structure of a Self-Assembly-Competent Form of a Hydrophobic Peptide Captured in a Soluble β-Sheet Scaffold.
J.Mol.Biol., 378, 2008
|
|
2OL7
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-01-18 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
2OL6
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-01-18 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
2G8C
 
 | |
2OL8
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-01-18 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
2LIE
 
 | | NMR structure of the lectin CCL2 | | Descriptor: | CCL2 lectin | | Authors: | Schubert, M, Walti, M.A, Egloff, P, Bleuler-Martinez, S, Aebi, M, Allain, F.F.-H, Kunzler, M. | | Deposit date: | 2011-08-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2OY1
 
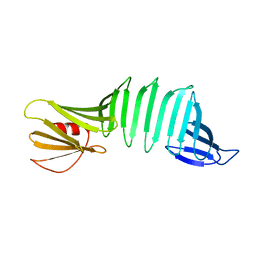 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-02-21 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
