6FDW
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-356 | | Descriptor: | (4aS,8aR)-2-(1-{2-aminothieno[2,3-d]pyrimidin-4-yl}piperidin-4-yl)-4-(3,4- dimethoxyphenyl)-1,2,4a,5,8,8a-hexahydrophthalazin-1-one, GLYCEROL, GUANIDINE, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | TbrPDEB1 structure with inhibitor NPD-356
To be published
|
|
6FE3
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-1439 | | Descriptor: | 1-(2-{4-[(4aS,8aR)-4-[3,4-bis(difluoromethoxy)phenyl]-1-oxo-1,2,4a,5,8,8a-hexahydrophthalazin-2-yl]piperidin-1-yl}-2-oxoethyl)-4,4-dimethylpiperidine-2,6-dione, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | TbrPDEB1 structure with inhibitor NPD-1439
To be published
|
|
6FW3
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-007 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{4-[(4aS,8aR)-4-(3,4-dimethoxyphenyl)-1-oxo-1,2,4a,5,8,8a-hexahydrophthalazin-2-yl]piperidin-1-yl}-2-oxoethyl)pyrrolidine-2,5-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-03-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-007
To be published
|
|
6FV9
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-007 | | Descriptor: | 1-(2-{4-[(4aS,8aR)-4-(3,4-dimethoxyphenyl)-1-oxo-1,2,4a,5,8,8a-hexahydrophthalazin-2-yl]piperidin-1-yl}-2-oxoethyl)pyrrolidine-2,5-dione, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | TbrPDEB1 structure with inhibitor NPD-007
To be published
|
|
6FVR
 
 | |
6FVS
 
 | |
6GNK
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form bound to Carba-NAD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 14-3-3 protein beta/alpha, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
4EFU
 
 | | Hsp90 Alpha N-terminal Domain in Complex with an Inhibitor 6-Hydroxy-3-(3-methyl-benzyl)-1H-indazole-5-carboxylic acid benzyl-methyl-amide | | Descriptor: | Heat shock protein HSP 90-alpha, N-benzyl-6-hydroxy-N-methyl-3-(3-methylbenzyl)-1H-indazole-5-carboxamide, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Graedler, U, Buchstaller, H.-P. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based discovery of hydroxy-indazole-carboxamides as novel small molecule inhibitors of Hsp90
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2O98
 
 | | Structure of the 14-3-3 / H+-ATPase plant complex | | Descriptor: | 14-3-3-like protein C, FUSICOCCIN, Plasma membrane H+ ATPase, ... | | Authors: | Ottmann, C, Weyand, M, Wittinghofer, A, Oecking, C. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+ -ATPase by combining X-ray crystallography and electron cryomicroscopy
Mol.Cell, 25, 2007
|
|
6FE7
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-356 | | Descriptor: | (4aS,8aR)-2-(1-{2-aminothieno[2,3-d]pyrimidin-4-yl}piperidin-4-yl)-4-(3,4- dimethoxyphenyl)-1,2,4a,5,8,8a-hexahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-29 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-356
To be published
|
|
6FET
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1439 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{4-[(4aS,8aR)-4-[3,4-bis(difluoromethoxy)phenyl]-1-oxo-1,2,4a,5,8,8a-hexahydrophthalazin-2-yl]piperidin-1-yl}-2-oxoethyl)-4,4-dimethylpiperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-1439
To be published
|
|
6FL9
 
 | |
6FDS
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-226 | | Descriptor: | 1-[2-[4-[(4~{a}~{S},8~{a}~{R})-4-(3,4-dimethoxyphenyl)-1-oxidanylidene-4~{a},5,8,8~{a}-tetrahydrophthalazin-2-yl]piperi din-1-yl]-2-oxidanylidene-ethyl]-4,4-dimethyl-piperidine-2,6-dione, GLYCEROL, GUANIDINE, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | TbrPDEB1 structure with inhibitor NPD-226
To be published
|
|
6FLI
 
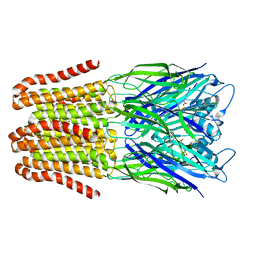 | |
6GNJ
 
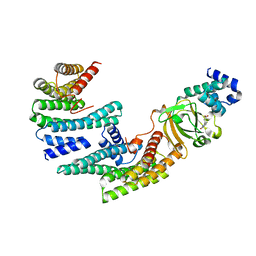 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form in complex with STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GN0
 
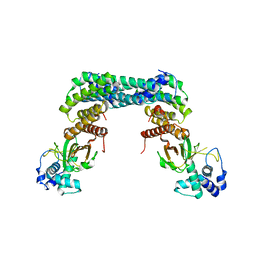 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form | | Descriptor: | 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GNN
 
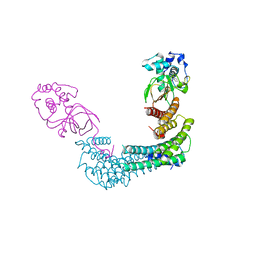 | | Exoenzyme T from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form bound to STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme T | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GN8
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6FVQ
 
 | |
2WHG
 
 | | Crystal Structure of the Di-Zinc Metallo-beta-lactamase VIM-4 from Pseudomonas aeruginosa | | Descriptor: | CITRATE ANION, VIM-4 METALLO-BETA-LACTAMASE, ZINC ION | | Authors: | Lassaux, P, Traore, D.A.K, Galleni, M, Ferrer, J.L. | | Deposit date: | 2009-05-05 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Characterization of the Subclass B1 Metallo-{Beta}-Lactamase Vim-4.
Antimicrob.Agents Chemother., 55, 2011
|
|
1N1X
 
 | | Crystal Structure Analysis of the monomeric [S-carboxyamidomethyl-Cys31, S-carboxyamidomethyl-Cys32] Bovine seminal ribonuclease | | Descriptor: | Ribonuclease, seminal | | Authors: | Sica, F, Di Fiore, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2002-10-21 | | Release date: | 2003-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The unswapped chain of bovine seminal ribonuclease: Crystal structure of the free and liganded monomeric derivative
Proteins, 52, 2003
|
|
1PNE
 
 | | CRYSTALLIZATION AND STRUCTURE DETERMINATION OF BOVINE PROFILIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | PROFILIN | | Authors: | Cedergren-Zeppezauer, E.S, Goonesekere, N.C.W, Rozycki, M.D, Myslik, J.C, Dauter, Z, Lindberg, U, Schutt, C.E. | | Deposit date: | 1995-05-05 | | Release date: | 1995-07-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and structure determination of bovine profilin at 2.0 A resolution.
J.Mol.Biol., 240, 1994
|
|
1S1N
 
 | | SH3 domain of human nephrocystin | | Descriptor: | Nephrocystin 1 | | Authors: | Le Maire, A, Weber, T, Saunier, S, Antignac, C, Ducruix, A, Dardel, F. | | Deposit date: | 2004-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SH3 domain of human nephrocystin and analysis of a mutation-causing juvenile nephronophthisis.
Proteins, 59, 2005
|
|
1QOB
 
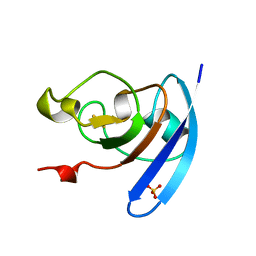 | | FERREDOXIN MUTATION D62K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1QOF
 
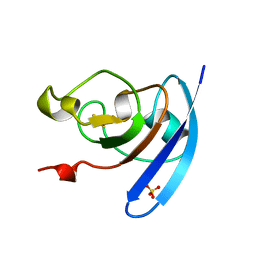 | | FERREDOXIN MUTATION Q70K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
