1KH0
 
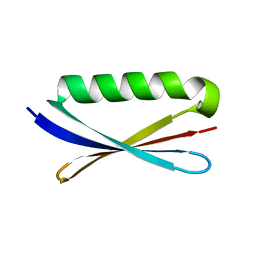 | | Accurate Computer Base Design of a New Backbone Conformation in the Second Turn of Protein L | | Descriptor: | protein L | | Authors: | O'Neill, J.W, Kuhlman, B, Kim, D.E, Zhang, K.Y, Baker, D. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Accurate computer-based design of a new backbone conformation in the second turn of protein L.
J.Mol.Biol., 315, 2002
|
|
5TX8
 
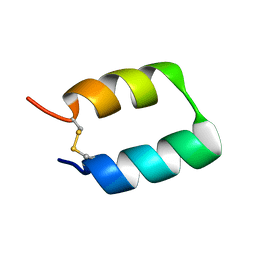 | |
5TRV
 
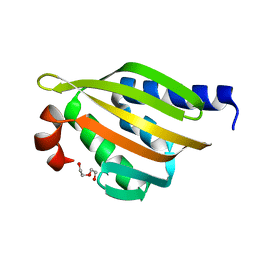 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TPJ
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TPH
 
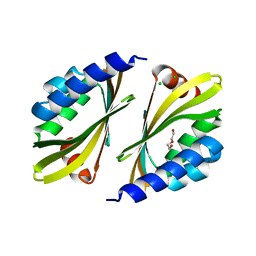 | | Crystal structure of a de novo designed protein homodimer with curved beta-sheet | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, de novo NTF2 homodimer | | Authors: | Basanta, B, Marcos, E, Oberdorfer, G, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TS4
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Chidyausiku, T.M, Marcos, E, Pereira, J.H, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
2KL8
 
 | | Solution NMR Structure of de novo designed ferredoxin-like fold protein, Northeast Structural Genomics Consortium Target OR15 | | Descriptor: | OR15 | | Authors: | Liu, G, Koga, N, Jiang, M, Koga, R, Xiao, R, Ciccosanti, C, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2KW5
 
 | | Solution NMR Structure of the Slr1183 protein from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Rossi, P, Forouhar, F, Lee, H, Lange, O, Mao, B, Lemak, A, Maglaqui, M, Belote, R, Ciccosanti, C, Foote, E, Sahdev, S, Acton, T, Xiao, R, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LCI
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, P-LOOP NTPASE FOLD, Northeast Structural Genomics Consortium Target OR36 (CASD Target) | | Descriptor: | Protein OR36 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Janjua, H, Ciccosanti, C, Lee, H, Acton, T.B, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR36
To be Published
|
|
2LR0
 
 | | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136 | | Descriptor: | P-loop ntpase fold | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-19 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136
To be Published
|
|
2LVB
 
 | | Solution NMR Structure DE NOVO DESIGNED PFK fold PROTEIN, Northeast Structural Genomics Consortium (NESG) Target OR250 | | Descriptor: | DE NOVO DESIGNED PFK fold PROTEIN | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2LRH
 
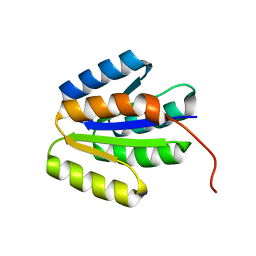 | | Solution NMR structure of de novo designed protein, p-loop ntpase fold, Northeast Structural Genomics Consortium target or137 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-30 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed protein, p-loop ntpase fold, Northeast Structural Genomics Consortium target or137
To be Published
|
|
2LN3
 
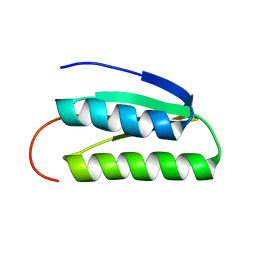 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, IF3-like fold, Northeast Structural Genomics Consortium Target OR135 (CASD target) | | Descriptor: | DE NOVO DESIGNED PROTEIN OR135 | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2LTA
 
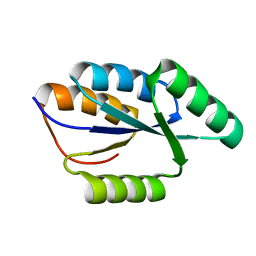 | | Solution NMR structure of De novo designed protein, rossmann 3x1 fold, Northeast Structural Genomics Consortium target OR157 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2MBL
 
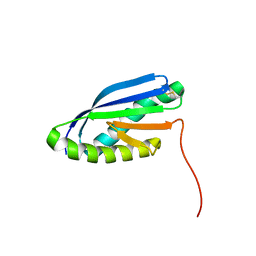 | | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33 | | Descriptor: | Top7 Fold Protein Top7m13 | | Authors: | Liu, G, Zanghellini, A.L, Chan, K, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-08-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33
To be Published
|
|
2MME
 
 | | Hybrid structure of the Shigella flexneri MxiH Type three secretion system needle | | Descriptor: | MxiH | | Authors: | Demers, J.P, Habenstein, B, Loquet, A, Vasa, S.K, Becker, S, Baker, D, Lange, A, Sgourakis, N.G. | | Deposit date: | 2014-03-14 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å), SOLID-STATE NMR | | Cite: | High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy.
Nat Commun, 5, 2014
|
|
2MR5
 
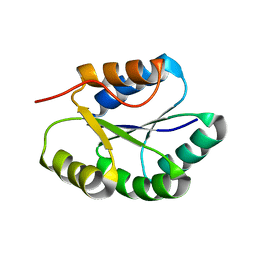 | | Solution NMR Structure of De novo designed Protein, Northeast Structural Genomics Consortium (NESG) Target OR457 | | Descriptor: | De novo designed Protein OR457 | | Authors: | Pulavarti, S, Nivon, L, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-01 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed Protein, Northeast Structural Genomics Consortium (NESG) Target OR457
To be Published
|
|
2LV8
 
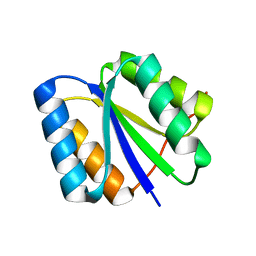 | | Solution NMR Structure de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium (NESG) Target OR16 | | Descriptor: | De novo designed rossmann 2x2 fold protein | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Ciccosanti, C, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2N3Z
 
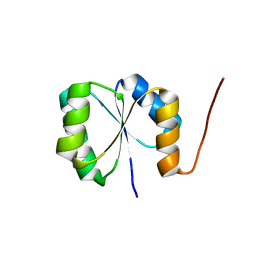 | | Solution NMR Structure of de novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | OR446 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-15 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
2MQ8
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-06-12 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Control over overall shape and size in de novo designed proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2N2T
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303 | | Descriptor: | OR303 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303
To be Published
|
|
2L15
 
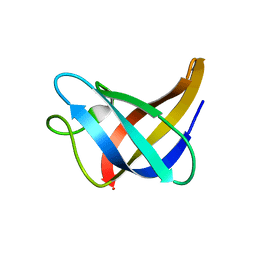 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | Descriptor: | Cold shock protein CspA | | Authors: | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
2LND
 
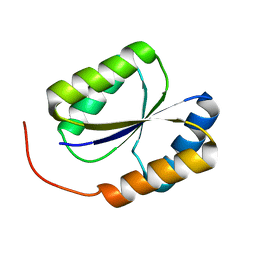 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, PFK fold, Northeast Structural Genomics Consortium Target OR134 | | Descriptor: | DE NOVO DESIGNED PROTEIN, PFK fold | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR134
To be Published
|
|
2N2U
 
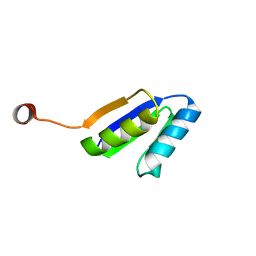 | | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358 | | Descriptor: | OR358 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358
To be Published
|
|
2N76
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414
To be Published
|
|
