6IBX
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 5 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-[(3~{S})-1-methylpiperidin-3-yl]pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6D2M
 
 | | Beta Carbonic anhydrase in complex with thiocyanate | | Descriptor: | Carbonic anhydrase, IMIDAZOLE, ZINC ION | | Authors: | Murray, A, Aggarwal, M, Pinard, M, McKenna, R. | | Deposit date: | 2018-04-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mapping of Anion Inhibitors to beta-Carbonic Anhydrase psCA3 from Pseudomonas aeruginosa.
ChemMedChem, 13, 2018
|
|
1G42
 
 | | STRUCTURE OF 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE (LINB) FROM SPHINGOMONAS PAUCIMOBILIS COMPLEXED WITH 1,2-DICHLOROPROPANE | | Descriptor: | 1,2-DICHLORO-PROPANE, 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, ACETATE ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-10-26 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
8K6T
 
 | | The minor pilin structure of FctB3 in Streptococcus | | Descriptor: | FctB3, GLYCEROL | | Authors: | Takebe, K, Sangawa, T, Suzuki, M, Nakata, M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of FctB3 crystal structure and insight into its structural stabilization and pilin linkage mechanisms.
Arch.Microbiol., 206, 2023
|
|
6NB3
 
 | | MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
1GDU
 
 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-ARG, SULFATE ION, TRYPSIN | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-29 | | Release date: | 2001-02-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6TLL
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4,5,6,7-TETRABROMOBENZOTRIAZOLE (tBBT) | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
8HUA
 
 | | Serial synchrotron crystallography structure of ba3-type cytochrome c oxidase from Thermus thermophilus using a goniometer compatible flow-cell | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Ghosh, S, Zoric, D, Bjelcic, M, Johannesson, J, Sandelin, E, Branden, G, Neutze, R. | | Deposit date: | 2022-12-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
6TLW
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4-BROMOBENZOTRIAZOLE | | Descriptor: | 7-bromanyl-1~{H}-benzotriazole, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
1GBQ
 
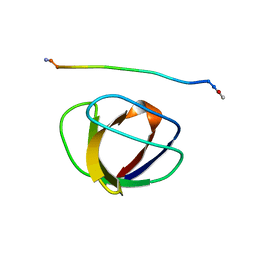 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1FY5
 
 | | Fusarium oxysporum trypsin at atomic resolution | | Descriptor: | GLY-ALA-LYS, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6NBE
 
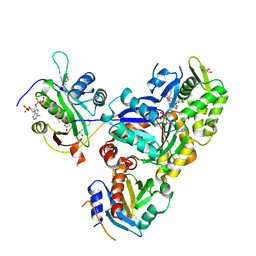 | | Ternary Complex of Ac-Alpha-Actin with Profilin and CoA-NAA80 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2018-12-07 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of actin N-terminal acetylation.
Sci Adv, 6, 2020
|
|
6NEE
 
 | |
1G5A
 
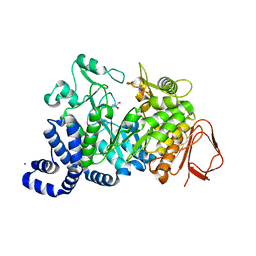 | | AMYLOSUCRASE FROM NEISSERIA POLYSACCHAREA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMYLOSUCRASE, ... | | Authors: | Skov, L.K, Mirza, O, Henriksen, A, De Montalk, G.P, Remaud-Simeon, M, Sarcabal, P, Willemot, R.-M, Monsan, P, Gajhede, M. | | Deposit date: | 2000-10-31 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Amylosucrase, A Glucan-synthesizing Enzyme from the alpha-Amylase Family
J.Biol.Chem., 276, 2001
|
|
6NFZ
 
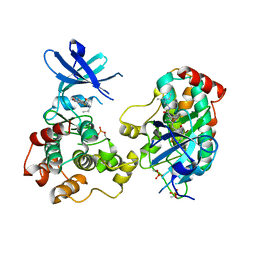 | | Crystal structure of diphosphorylated HPK1 kinase domain in complex with sunitinib in the active state. | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carboxamide | | Authors: | Johnson, E, McTigue, M, Cronin, C.N. | | Deposit date: | 2018-12-21 | | Release date: | 2019-05-01 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (2.966 Å) | | Cite: | Multiple conformational states of the HPK1 kinase domain in complex with sunitinib reveal the structural changes accompanying HPK1 trans-regulation.
J.Biol.Chem., 294, 2019
|
|
8H0M
 
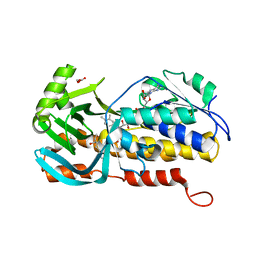 | | Crystal structure of VioD | | Descriptor: | (2S)-2-ethylhexan-1-ol, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Xu, M, Ran, T, Wang, W. | | Deposit date: | 2022-09-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
5H0Q
 
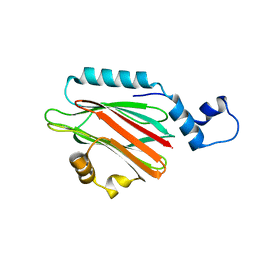 | | Crystal structure of lipid binding protein Nakanori at 1.5A | | Descriptor: | Lipid binding protein | | Authors: | Makino, A, Abe, M, Ishitsuka, R, Murate, M, Kishimoto, T, Sakai, S, Hullin-Matsuda, F, Shimada, Y, Inaba, T, Miyatake, H, Tanaka, H, Kurahashi, A, Pack, C.G, Kasai, R.S, Kubo, S, Schieber, N.L, Dohmae, N, Tochio, N, Hagiwara, K, Sasaki, Y, Aida, Y, Fujimori, F, Kigawa, T, Nishikori, K, Parton, R.G, Kusumi, A, Sako, Y, Anderluh, G, Yamashita, M, Kobayashi, T, Greimel, P, Kobayashi, T. | | Deposit date: | 2016-10-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C
FASEB J., 31, 2017
|
|
1GDN
 
 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-LYS, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6TLO
 
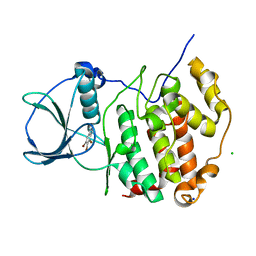 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4,5,6-TRIBROMOBENZOTRIAZOLE | | Descriptor: | 5,6,7-tris(bromanyl)-1~{H}-benzotriazole, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
7X93
 
 | |
7X95
 
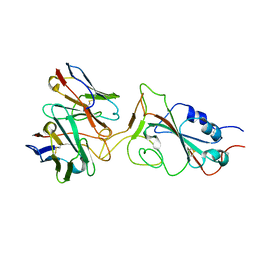 | |
6ZBA
 
 | | Crystal structure of PDE4D2 in complex with inhibitor LEO39652 | | Descriptor: | 1,2-ETHANEDIOL, 2-methylpropyl 1-[8-methoxy-5-(1-oxidanylidene-3~{H}-2-benzofuran-5-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]cyclopropane-1-carboxylate, DIMETHYL SULFOXIDE, ... | | Authors: | Akutsu, M, Hakansson, M, Welin, M, Svensson, A, Logan, D.T, Sorensen, M.D. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Early Clinical Development of Isobutyl 1-[8-Methoxy-5-(1-oxo-3 H -isobenzofuran-5-yl)-[1,2,4]triazolo[1,5- a ]pyridin-2-yl]cyclopropanecarboxylate (LEO 39652), a Novel "Dual-Soft" PDE4 Inhibitor for Topical Treatment of Atopic Dermatitis.
J.Med.Chem., 63, 2020
|
|
6TLV
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5-BROMOBENZOTRIAZOLE | | Descriptor: | 6-bromanyl-1~{H}-benzotriazole, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
7X94
 
 | |
6NSV
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | Descriptor: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2019-01-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
