7XIX
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNQ
 
 | | SARS-CoV-2 Omicron BA.4 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIY
 
 | | SARS-CoV-2 Omicron BA.3 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIW
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIZ
 
 | | SARS-CoV-2 Omicron BA.3 variant spike (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNR
 
 | | SARS-CoV-2 Omicron BA.2.13 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7Y0W
 
 | | Local structure of BD55-5514 and BD55-5840 Fab and Omicron BA.1 RBD complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-5514H, BD55-5514L, ... | | Authors: | Zhang, Z, Xiao, J. | | Deposit date: | 2022-06-06 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
5YYL
 
 | | Structure of Major Royal Jelly Protein 1 Oligomer | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tian, W, Chen, Z. | | Deposit date: | 2017-12-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architecture of the native major royal jelly protein 1 oligomer.
Nat Commun, 9, 2018
|
|
3OND
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ONF
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with cordycepin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3'-DEOXYADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6J1O
 
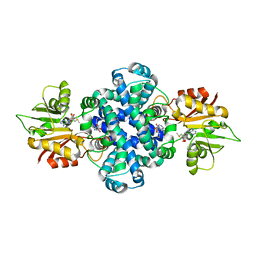 | |
6J46
 
 | | LepI-SAH complex structure | | Descriptor: | O-methyltransferase lepI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
8DKC
 
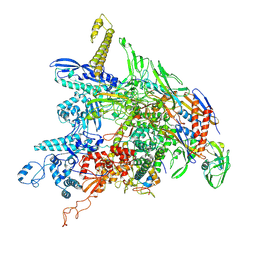 | | P. gingivalis RNA Polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Bu, F. | | Deposit date: | 2022-07-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structure of Porphyromonas gingivalis RNA Polymerase.
J.Mol.Biol., 436, 2024
|
|
8YZR
 
 | |
8YZZ
 
 | | The structure of HLA-A*2402 complex with peptide from SARS-CoV-2 S448-456 NYNYLYRLF(Prototype) | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1 | | Authors: | Shang, B.L, Zhang, J.N, Tian, J.M, Liu, J. | | Deposit date: | 2024-04-08 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots.
Nat.Immunol., 26, 2025
|
|
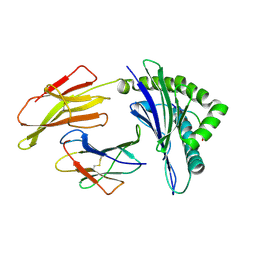
8Z07
 
 | |
8Z06
 
 | |
8Z05
 
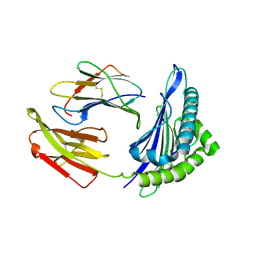 | | The structure of HLA-A*0201 complex with peptide from SARS-CoV-2 N222-230 LLLDRLNKL(BA.2.86/JN.1) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Zhang, J.N, Tian, J.M, Liu, J. | | Deposit date: | 2024-04-09 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots.
Nat.Immunol., 26, 2025
|
|
8GTY
 
 | |
8Z08
 
 | |
8YZW
 
 | |
6J24
 
 | | Crystal structure of a SAM-dependent methyltransferase LepI in complex with its substrate | | Descriptor: | (3~{S},4'~{R},4'~{a}~{S},6'~{R},8'~{a}~{S})-4',6'-dimethyl-5-phenyl-spiro[1~{H}-pyridine-3,5'-2,3,4,4~{a},6,8~{a}-hexahydro-1~{H}-naphthalene]-2,4-dione, O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2018-12-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
7LC5
 
 | |
4JDZ
 
 | |
8K31
 
 | | The complex of WRKY33 C terminal DBD and SIB1 | | Descriptor: | Probable WRKY transcription factor 33, Sigma factor binding protein 1, chloroplastic, ... | | Authors: | Dong, X, Gong, Z, Hu, Y.F. | | Deposit date: | 2023-07-14 | | Release date: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1.
Commun Biol, 7, 2024
|
|
