1ICQ
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH 9R,13R-OPDA | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, 9R,13R-12-OXOPHYTODIENOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1FIU
 
 | | TETRAMERIC RESTRICTION ENDONUCLEASE NGOMIV IN COMPLEX WITH CLEAVED DNA | | Descriptor: | ACETIC ACID, DNA (5'-D(*TP*GP*CP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*CP*GP*C)-3'), ... | | Authors: | Deibert, M, Grazulis, S, Sasnauskas, G, Siksnys, V, Huber, R. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA.
Nat.Struct.Biol., 7, 2000
|
|
1FS7
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1R6V
 
 | | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin | | Descriptor: | CALCIUM ION, subtilisin-like serine protease | | Authors: | Kim, J.S, Kluskens, L.D, de Vos, W.M, Huber, R, van der Oost, J. | | Deposit date: | 2003-10-17 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin.
J.Mol.Biol., 335, 2004
|
|
1ICP
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH PEG400 | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1KFX
 
 | | Crystal Structure of Human m-Calpain Form I | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KL7
 
 | | Crystal Structure of Threonine Synthase from Yeast | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Threonine Synthase | | Authors: | Garrido-Franco, M, Ehlert, S, Messerschmidt, A, Marinkovic, S, Huber, R, Laber, B, Bourenkov, G.P, Clausen, T. | | Deposit date: | 2001-12-11 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of threonine synthase from yeast.
J.Biol.Chem., 277, 2002
|
|
1U80
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CMP complex | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHATE ION | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1KFU
 
 | | Crystal Structure of Human m-Calpain Form II | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FS9
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES-AZIDE COMPLEX | | Descriptor: | AZIDE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1KLJ
 
 | | Crystal structure of uninhibited factor VIIa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | Authors: | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
2IMN
 
 | |
1FB1
 
 | | CRYSTAL STRUCTURE OF HUMAN GTP CYCLOHYDROLASE I | | Descriptor: | GTP CYCLOHYDROLASE I, ISOPROPYL ALCOHOL, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, G, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-14 | | Release date: | 2000-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1U7W
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CTP-complex | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1DD4
 
 | | Crystal structure of ribosomal protein l12 from thermotoga maritim | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12, HEXATANTALUM DODECABROMIDE | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
Embo J., 19, 2000
|
|
1FBX
 
 | | CRYSTAL STRUCTURE OF ZINC-CONTAINING E.COLI GTP CYCLOHYDROLASE I | | Descriptor: | CHLORIDE ION, GTP CYCLOHYDROLASE I, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, A, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-17 | | Release date: | 2001-02-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ELQ
 
 | | CRYSTAL STRUCTURE OF THE CYSTINE C-S LYASE C-DES | | Descriptor: | L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KZ6
 
 | | Mutant enzyme W63Y/L119F Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1G8F
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1KYY
 
 | | Lumazine Synthase from S.pombe bound to nitropyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1T1G
 
 | | High Resolution Crystal Structure of Mutant E23A of Kumamolisin, a sedolisin type proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1KZ1
 
 | | Mutant enzyme W27G Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1PYT
 
 | | TERNARY COMPLEX OF PROCARBOXYPEPTIDASE A, PROPROTEINASE E, AND CHYMOTRYPSINOGEN C | | Descriptor: | CALCIUM ION, CHYMOTRYPSINOGEN C, PROCARBOXYPEPTIDASE A, ... | | Authors: | Gomis-Ruth, F.X, Gomez, M, Bode, W, Huber, R, Aviles, F.X. | | Deposit date: | 1995-06-21 | | Release date: | 1997-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The three-dimensional structure of the native ternary complex of bovine pancreatic procarboxypeptidase A with proproteinase E and chymotrypsinogen C.
EMBO J., 14, 1995
|
|
1T1E
 
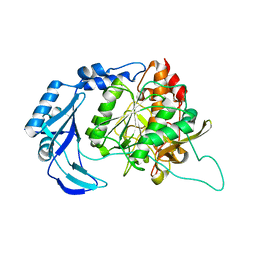 | | High Resolution Crystal Structure of the Intact Pro-Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
2FCB
 
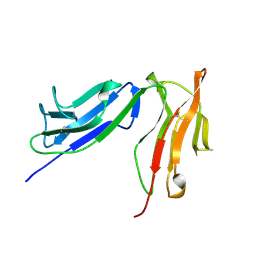 | | HUMAN FC GAMMA RECEPTOR IIB ECTODOMAIN (CD32) | | Descriptor: | PROTEIN (FC GAMMA RIIB) | | Authors: | Sondermann, P, Huber, R, Jacob, U. | | Deposit date: | 1999-01-07 | | Release date: | 2000-03-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the soluble form of the human fcgamma-receptor IIb: a new member of the immunoglobulin superfamily at 1.7 A resolution.
EMBO J., 18, 1999
|
|
