7NWA
 
 | |
7NYA
 
 | |
7NWM
 
 | |
7NWC
 
 | |
7NXN
 
 | |
7NY2
 
 | |
7OV0
 
 | |
1HDL
 
 | | LEKTI domain one | | Descriptor: | SERINE PROTEINASE INHIBITOR LEKTI | | Authors: | Lauber, T, Roesch, P, Marx, U.C. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Homologous Proteins with Different Folds: The Three-Dimensional Structures of Domains 1 and 6 of the Multiple Kazal-Type Inhibitor Lekti
J.Mol.Biol., 328, 2003
|
|
1H0Z
 
 | | LEKTI domain six | | Descriptor: | SERINE PROTEASE INHIBITOR KAZAL-TYPE 5, CONTAINS HEMOFILTRATE PEPTIDE HF6478, HEMOFILTRATE PEPTIDE HF7665 | | Authors: | Lauber, T, Roesch, P, Marx, U.C. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Homologous Proteins with Different Folds: The Three-Dimensional Structures of Domains 1 and 6 of the Multiple Kazal-Type Inhibitor Lekti
J.Mol.Biol., 328, 2003
|
|
7Q1H
 
 | |
5M96
 
 | |
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
8B0R
 
 | | Structure of the CalpL/cA4 complex | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), SMODS-associated and fused to various effectors domain-containing protein, SULFATE ION, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8B0U
 
 | | Structure of the CalpL/T10 complex | | Descriptor: | CalpT10, GLYCEROL, SAVED domain-containing protein, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
3TPK
 
 | | Crystal structure of the oligomer-specific KW1 antibody fragment | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, Immunoglobulin heavy chain antibody variable domain KW1 | | Authors: | Parthier, C, Morgado, I, Stubbs, M.T, Fandrich, M. | | Deposit date: | 2011-09-08 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of beta-amyloid oligomer recognition with a conformational antibody fragment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5HBJ
 
 | | CDK8-CYCC IN COMPLEX WITH 8-[2-Amino-3-chloro-5-(1-methyl-1H-indazol-5-yl)-pyridin-4-yl]-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 1,2-ETHANEDIOL, 8-[2-azanyl-3-chloranyl-5-(1-methylindazol-5-yl)pyridin-4-yl]-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MBW
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH Pep#3 | | Descriptor: | BACE1 INHIBITOR PEPTIDE Pep#3, Beta-secretase 1, CHLORIDE ION | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-09 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MCO
 
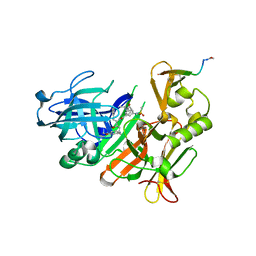 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE INHIBITOR GRL-8234 AND EXOSITE PEPTIDE | | Descriptor: | BACE-1 EXOSITE PEPTIDE, Beta-secretase 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
7KDL
 
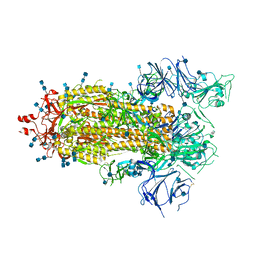 | |
7KE9
 
 | |
7KE6
 
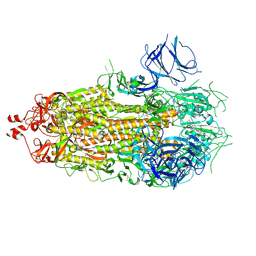 | |
7KEB
 
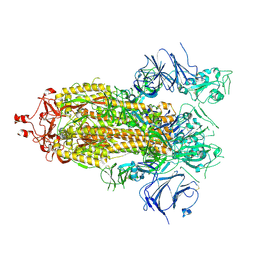 | |
7KDI
 
 | |
7KDH
 
 | |
