5TTR
 
 | | LEU 55 PRO TRANSTHYRETIN CRYSTAL STRUCTURE | | Descriptor: | TRANSTHYRETIN | | Authors: | Sebastiao, M.P, Saraiva, M.J, Damas, A.M. | | Deposit date: | 1998-04-30 | | Release date: | 1999-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of amyloidogenic Leu55 --> Pro transthyretin variant reveals a possible pathway for transthyretin polymerization into amyloid fibrils.
J.Biol.Chem., 273, 1998
|
|
4EKF
 
 | |
1EFI
 
 | |
353D
 
 | | CRYSTAL STRUCTURE OF DOMAIN A OF THERMUS FLAVUS 5S RRNA AND THE CONTRIBUTION OF WATER MOLECULES TO ITS STRUCTURE | | Descriptor: | RNA (5'-R(*AP*UP*CP*CP*CP*CP*CP*GP*UP*GP*CP*C)-3'), RNA (5'-R(*GP*GP*UP*GP*CP*GP*GP*GP*GP*GP*AP*U)-3') | | Authors: | Betzel, C, Lorenz, S, Furste, J.P, Bald, R, Zhang, M, Schneider, T.R, Wilson, K.S, Erdmann, V.A. | | Deposit date: | 1997-09-29 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of domain A of Thermus flavus 5S rRNA and the contribution of water molecules to its structure.
FEBS Lett., 351, 1994
|
|
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AFQ
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form II) | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
2PSP
 
 | |
6MT1
 
 | |
6MT2
 
 | |
1FXW
 
 | |
3NUL
 
 | | Profilin I from Arabidopsis thaliana | | Descriptor: | GLYCEROL, PROFILIN I, SULFATE ION | | Authors: | Thorn, K, Christensen, H.E.M, Shigeta, R, Huddler, D, Chua, N.-H, Shalaby, L, Lindberg, U, Schutt, C.E. | | Deposit date: | 1996-11-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a major allergen from plants.
Structure, 5, 1997
|
|
5W19
 
 | | Tryptophan indole-lyase complex with oxindolyl-L-alanine | | Descriptor: | 1-carboxy-1-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]azaniumyl}-2-[(3R)-2-oxo-2,3-dihydro-1H-indol-3-yl]ethan-1-ide, POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1X1V
 
 | | Structure Of Banana Lectin- Methyl-Alpha-Mannose Complex | | Descriptor: | HEXANE-1,6-DIOL, ZINC ION, lectin, ... | | Authors: | Singh, D.D, Saikrishnan, K, Kumar, P, Surolia, A, Sekar, K, Vijayan, M. | | Deposit date: | 2005-04-14 | | Release date: | 2005-11-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unusual sugar specificity of banana lectin from Musa paradisiaca and its probable evolutionary origin. Crystallographic and modelling studies
Glycobiology, 15, 2005
|
|
1A2I
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | Deposit date: | 1998-01-05 | | Release date: | 1998-07-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
1A0K
 
 | |
1B0D
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | LYSOZYME, PARA-TOLUENE SULFONATE | | Authors: | Vaney, M.C, Broutin, I, Retailleau, P, Lafont, S, Hamiaux, C, Prange, T, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-07 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1B44
 
 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | PROTEIN (B-POL SUBUNIT OF HEAT-LABILE ENTEROTOXIN) | | Authors: | Matkovic-Calogovic, D, Loregian, A, D'Acunto, M.R, Battistutta, R, Tossi, A, Palu, G, Zanotti, G. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
1WAP
 
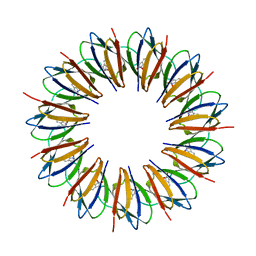 | |
1UPD
 
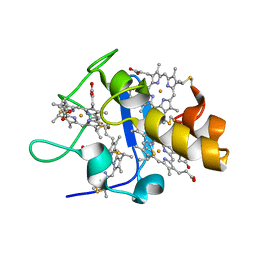 | | Oxidized STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
1B2K
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | IODIDE ION, PROTEIN (LYSOZYME) | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-26 | | Release date: | 1998-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1UP9
 
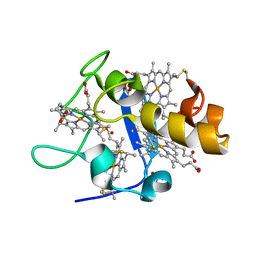 | | REDUCED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
1C54
 
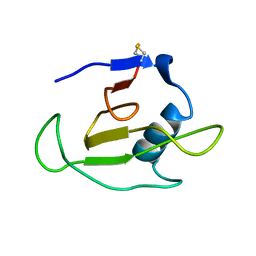 | | SOLUTION STRUCTURE OF RIBONUCLEASE SA | | Descriptor: | RIBONUCLEASE SA | | Authors: | Laurents, D.V, Canadillas-Perez, J.M, Santoro, J, Schell, D, Pace, C.N, Rico, M, Bruix, M. | | Deposit date: | 1999-10-22 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ribonuclease Sa.
Proteins, 44, 2001
|
|
1C7S
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT D539A COMPLEXED WITH DI-N-ACETYL-BETA-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-14 | | Release date: | 2000-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
