3MZV
 
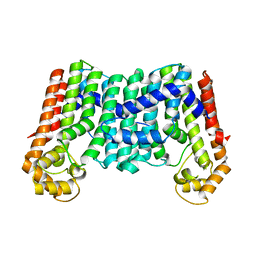 | | Crystal structure of a decaprenyl diphosphate synthase from Rhodobacter capsulatus | | Descriptor: | Decaprenyl diphosphate synthase | | Authors: | Quartararo, C.E, Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5XYF
 
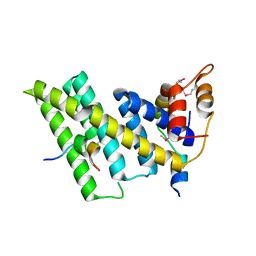 | | Crystal structure of the human TIN2-TPP1-TRF2 telomeric complex | | Descriptor: | Adrenocortical dysplasia protein homolog, TERF1-interacting nuclear factor 2, Telomeric repeat-binding factor 2 | | Authors: | Hu, C, Chen, Y. | | Deposit date: | 2017-07-07 | | Release date: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural and functional analyses of the mammalian TIN2-TPP1-TRF2 telomeric complex.
Cell Res., 27, 2017
|
|
8DIK
 
 | |
5WQD
 
 | |
7E80
 
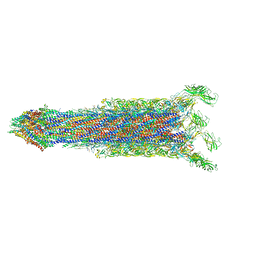 | | Cryo-EM structure of the flagellar rod with hook and export apparatus from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E81
 
 | | Cryo-EM structure of the flagellar MS ring with FlgB-Dc loop and FliE-helix 1 from Salmonella | | Descriptor: | Flagellar M-ring protein, FlgB-Dc loop, FliE helix 1 | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E82
 
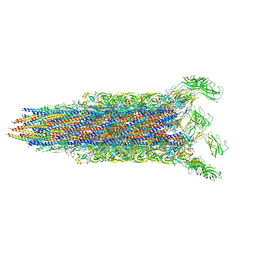 | | Cryo-EM structure of the flagellar rod with partial hook from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
3N75
 
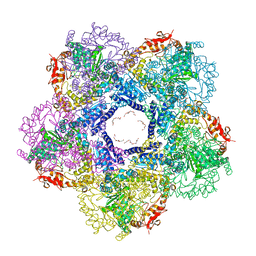 | | X-ray Crystal Structure of the Escherichia coli Inducible Lysine Decarboxylase LdcI | | Descriptor: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, HEXAETHYLENE GLYCOL, ... | | Authors: | Kanjee, U, Alexopoulos, E, Pai, E.F, Houry, W.A. | | Deposit date: | 2010-05-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase.
Embo J., 30, 2011
|
|
3Q16
 
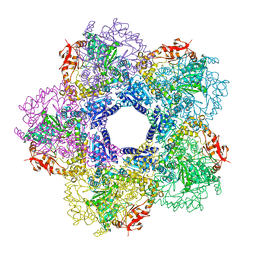 | |
2L3N
 
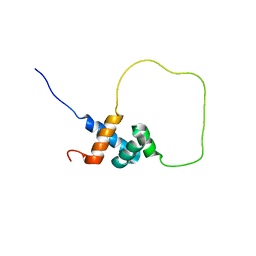 | | Solution structure of Rap1-Taz1 fusion protein | | Descriptor: | DNA-binding protein rap1,Telomere length regulator taz1 | | Authors: | Zhou, Z.R, Wang, F, Chen, Y, Lei, M, Hu, H. | | Deposit date: | 2010-09-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7MEM
 
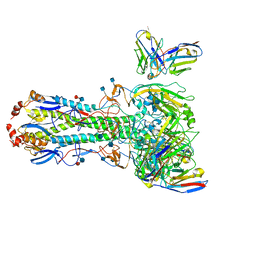 | |
1A19
 
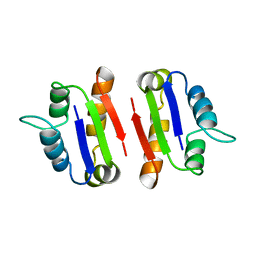 | | BARSTAR (FREE), C82A MUTANT | | Descriptor: | BARSTAR | | Authors: | Ratnaparkhi, G.S, Varadarajan, R. | | Deposit date: | 1997-12-25 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discrepancies between the NMR and X-ray structures of uncomplexed barstar: analysis suggests that packing densities of protein structures determined by NMR are unreliable.
Biochemistry, 37, 1998
|
|
7DXJ
 
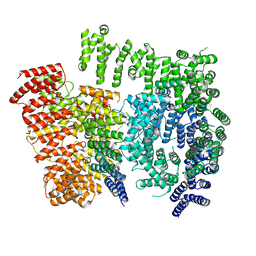 | |
7DXK
 
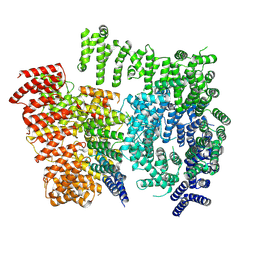 | |
6KAF
 
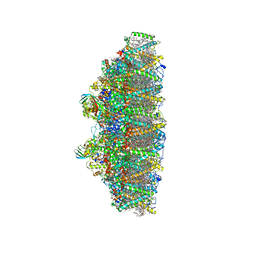 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4LR6
 
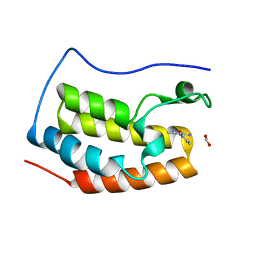 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | Descriptor: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
1WMA
 
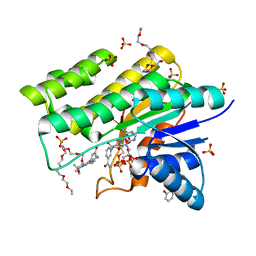 | | Crystal structure of human CBR1 in complex with Hydroxy-PP | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, ... | | Authors: | Rauh, D, Bateman, R, Shokat, K.M. | | Deposit date: | 2004-07-06 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An unbiased cell morphology-based screen for new, biologically active small molecules
Plos Biol., 3, 2005
|
|
3NUL
 
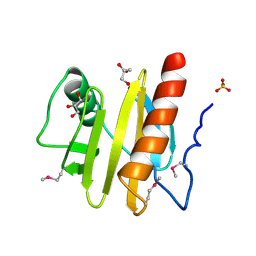 | | Profilin I from Arabidopsis thaliana | | Descriptor: | GLYCEROL, PROFILIN I, SULFATE ION | | Authors: | Thorn, K, Christensen, H.E.M, Shigeta, R, Huddler, D, Chua, N.-H, Shalaby, L, Lindberg, U, Schutt, C.E. | | Deposit date: | 1996-11-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a major allergen from plants.
Structure, 5, 1997
|
|
6JNX
 
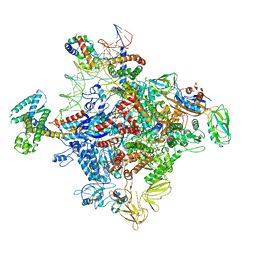 | | Cryo-EM structure of a Q-engaged arrested complex | | Descriptor: | Antiterminator Q protein, DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Feng, Y, Shi, J. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of Q-dependent transcription antitermination.
Nat Commun, 10, 2019
|
|
6JNY
 
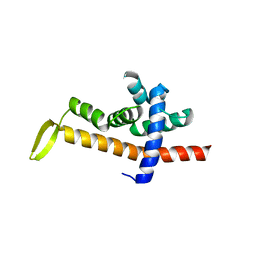 | |
1D62
 
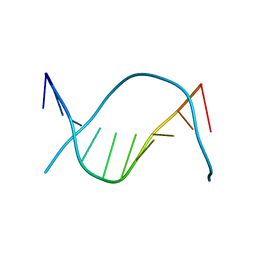 | |
1A0K
 
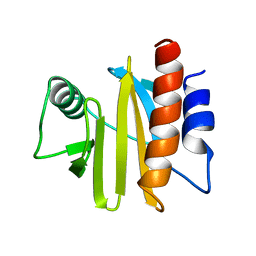 | |
1D61
 
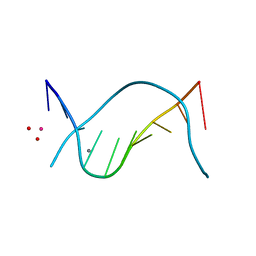 | | THE STRUCTURE OF THE B-DNA DECAMER C-C-A-A-C-I-T-T-G-G: MONOCLINIC FORM | | Descriptor: | CACODYLATE ION, CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*CP*IP*TP*TP*GP*G)-3') | | Authors: | Lipanov, A, Kopka, M.L, Kaczor-Grzeskowiak, M, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-02-26 | | Release date: | 1993-04-15 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA.
Biochemistry, 32, 1993
|
|
2NQ0
 
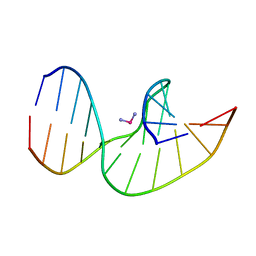 | | Solution Structures of a DNA Dodecamer Duplex with a Cisplatin 1,2-d(GG) Intrastrand Cross-Link | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', Cisplatin | | Authors: | Wu, Y, Bhattacharyya, D, Chaney, S, Campbell, S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a DNA Dodecamer Duplex with and without a Cisplatin 1,2-d(GG) Intrastrand Cross-Link: Comparison with the Same DNA Duplex Containing an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link
Biochemistry, 46, 2007
|
|
2NQ4
 
 | | Solution Structures of a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3' | | Authors: | Bhattacharyya, D, Wu, Y, Chaney, S, Campbell, S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a DNA Dodecamer Duplex with and without a Cisplatin 1,2-d(GG) Intrastrand Cross-Link: Comparison with the Same DNA Duplex Containing an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link
Biochemistry, 46, 2007
|
|
