3F7M
 
 | | Crystal structure of apo Cuticle-Degrading Protease (ver112) from Verticillium psalliotae | | Descriptor: | Alkaline serine protease ver112 | | Authors: | Liang, L, Lou, Z, Ye, F, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-09 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
4IB4
 
 | | Crystal structure of the chimeric protein of 5-HT2B-BRIL in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, ... | | Authors: | Wacker, D, Wang, C, Katritch, V, Han, G.W, Huang, X, Vardy, E, McCorvy, J.D, Jiang, Y, Chu, M, Siu, F.Y, Liu, W, Xu, H.E, Cherezov, V, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural features for functional selectivity at serotonin receptors.
Science, 340, 2013
|
|
2OF2
 
 | | crystal structure of furanopyrimidine 8 bound to lck | | Descriptor: | 2,3-DIPHENYL-N-(2-PIPERAZIN-1-YLETHYL)FURO[2,3-B]PYRIDIN-4-AMINE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Martin, M.W. | | Deposit date: | 2007-01-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel 2,3-diarylfuro[2,3-b]pyridin-4-amines as potent and selective inhibitors of Lck: Synthesis, SAR, and pharmacokinetic properties.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3G6L
 
 | |
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | Descriptor: | CAFFEINE, Chitinase | | Authors: | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | Deposit date: | 2009-02-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
3DA6
 
 | | Crystal Structure of human JNK3 complexed with N-(3-methyl-4-(3-(2-(methylamino)pyrimidin-4-yl)pyridin-2-yloxy)naphthalen-1-yl)-1H-benzo[d]imidazol-2-amine | | Descriptor: | Mitogen-activated protein kinase 10, N-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-1H-benzimidazol-2-amine | | Authors: | Cee, V.J, Cheng, A.C, Romero, K, Bellon, S, Mohr, C, Whittington, D.A, Bready, J, Caenepeel, S, Coxon, A, Deak, H.L, Hodous, B.L, Kim, J.L, Lin, J, Nguyen, H, Olivieri, P.R, Patel, V.F, Wang, L, Hughes, P, Geuns-Meyer, S. | | Deposit date: | 2008-05-28 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4FA6
 
 | |
3HBW
 
 | |
7E3O
 
 | |
6LDZ
 
 | | Crystal structure of Rv0222 from Mycobacterium tuberculosis | | Descriptor: | Probable enoyl-CoA hydratase EchA1 (Enoyl hydrase) (Unsaturated acyl-CoA hydratase) (Crotonase) | | Authors: | Li, J, Ran, Y.J, Wang, L, Wu, J.H, Ge, B.X, Rao, Z.H. | | Deposit date: | 2019-11-23 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Host-mediated ubiquitination of a mycobacterial protein suppresses immunity.
Nature, 577, 2020
|
|
5ZYA
 
 | | SF3b spliceosomal complex bound to E7107 | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Finci, L.I, Larsen, N.A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
7DTD
 
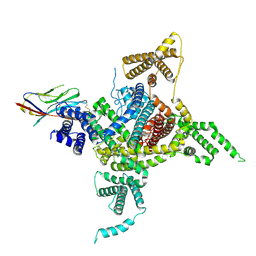 | | Voltage-gated sodium channel Nav1.1 and beta4 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 1 subunit alpha, ... | | Authors: | Yan, N, Pan, X, Li, Z, Huang, G. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Comparative structural analysis of human Na v 1.1 and Na v 1.5 reveals mutational hotspots for sodium channelopathies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5GMJ
 
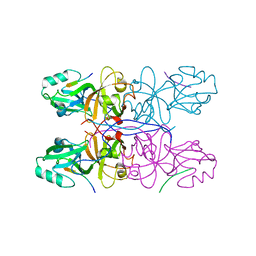 | | Crystal Structure of GRASP55 GRASP domain in complex with JAM-B C-terminus | | Descriptor: | Golgi reassembly-stacking protein 2, Junctional adhesion molecule B | | Authors: | Shi, N, Shi, X, Morelli, X, Betzi, S, Huang, X. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.986 Å) | | Cite: | Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis
PLoS Genet., 13, 2017
|
|
5YWO
 
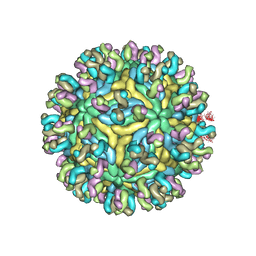 | | Structure of JEV-2F2 Fab complex | | Descriptor: | 2F2 heavy chain, 2F2 light chain, JEV E protein, ... | | Authors: | Qiu, X, Lei, Y.F, Yang, P, Gao, Q, Wang, N, Cao, L, Yuan, S, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2018-09-12 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5GMI
 
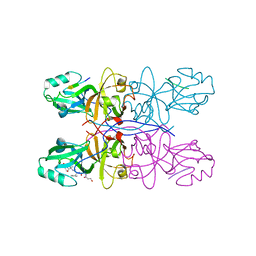 | | Crystal Structure of GRASP55 GRASP domain in complex with JAM-C C-terminus | | Descriptor: | Golgi reassembly-stacking protein 2, Junctional adhesion molecule C | | Authors: | Shi, N, Shi, X, Morelli, X, Betzi, S, Huang, X. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis
PLoS Genet., 13, 2017
|
|
5GML
 
 | |
5YWF
 
 | | Crystal structure of 2H4 Fab | | Descriptor: | 2H4 heavy chain, 2H4 light chain | | Authors: | Qiu, X, Lei, Y, Yang, P, Gao, Q, Wang, N, Cao, L, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
8HIF
 
 | | One asymmetric unit of Singapore grouper iridovirus capsid | | Descriptor: | Major capsid protein, Penton protein (VP14), VP137, ... | | Authors: | Zhao, Z.N, Liu, C.C, Zhu, D.J, Qi, J.X, Zhang, X.Z, Gao, G.F. | | Deposit date: | 2022-11-20 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic architecture of Singapore grouper iridovirus and implications for giant virus assembly.
Nat Commun, 14, 2023
|
|
5INW
 
 | | Structure of reaction loop cleaved lamprey angiotensinogen | | Descriptor: | C-terminal peptide of Putative angiotensinogen, Putative angiotensinogen, SULFATE ION | | Authors: | Wei, H, Zhou, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heparin Binds Lamprey Angiotensinogen and Promotes Thrombin Inhibition through a Template Mechanism
J.Biol.Chem., 291, 2016
|
|
5YWP
 
 | | JEV-2H4 Fab complex | | Descriptor: | 2H4 heavy chain, 2H4 light chain, JEV E protein, ... | | Authors: | Qiu, X.D, Lei, Y.F, Yang, P, Gao, Q, WANG, N, Cao, L, Yuan, S, Wang, X.X, Xu, Z.K, Rao, Z.H. | | Deposit date: | 2017-11-29 | | Release date: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5GY3
 
 | |
7TGE
 
 | |
7TOU
 
 | |
7TOV
 
 | |
7TOX
 
 | |
