6A57
 
 | | Structure of histone demethylase REF6 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*AP*A)-3'), GLYCEROL, ... | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
7W1V
 
 | | Active state CI from Rotenone-NADH dataset, Subclass 1 | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-20 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6J2W
 
 | | The structure of OBA3-OTA complex | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*CP*GP*AP*AP*GP*CP*GP*GP*GP*TP*CP*CP*CP*G)-3'), N-[(3R)-5-chloro-8-hydroxy-3-methyl-1-oxo-3,4-dihydro-1H-2-benzopyran-7-carbonyl]-D-phenylalanine | | Authors: | Xu, G.H, Li, C.G. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-guided post-SELEX optimization of an ochratoxin A aptamer.
Nucleic Acids Res., 47, 2019
|
|
4R8Q
 
 | |
5Y2G
 
 | | Structure of MBP tagged GBS CAMP | | Descriptor: | Maltose-binding periplasmic protein,Protein B, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2017-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of the CAMP factor of Streptococcus agalactiae with the aid of an MBP tag and insights into membrane-surface attachment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8V7O
 
 | |
8WJQ
 
 | | Cryo-EM structure of URAT1(R477S)-Urate complex | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Qian, H.W, He, J.J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
8WJH
 
 | | Cryo-EM structure of OAT4 | | Descriptor: | CHLORIDE ION, Solute carrier family 22 member 11 | | Authors: | Qian, H.W, He, J.J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
8WJG
 
 | | Cryo-EM structure of URAT1(R477S) | | Descriptor: | Solute carrier family 22 member 12 | | Authors: | Qian, H.W, He, J.J. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | Descriptor: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | Zhao, S, Huang, J, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
1HCF
 
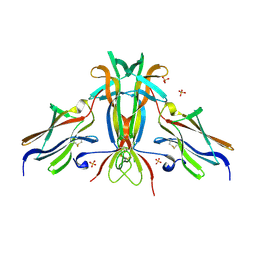 | | Crystal structure of TrkB-d5 bound to neurotrophin-4/5 | | Descriptor: | BDNF/NT-3 GROWTH FACTORS RECEPTOR, NEUROTROPHIN-4, SULFATE ION | | Authors: | Banfield, M.J, Naylor, R.L, Robertson, A.G.S, Allen, S.J, Dawbarn, D, Brady, R.L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity in Trk-Receptor:Neurotrophin Interaction: The Crystal Structure of Trkb-D5 in Complex with Neurotrophin-4/5
Structure, 9, 2001
|
|
6JDS
 
 | |
6J9O
 
 | |
6JDU
 
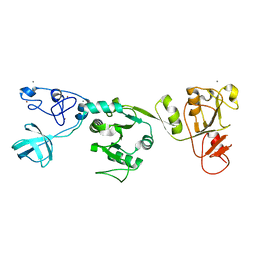 | | Crystal structure of PRRSV nsp10 (helicase) | | Descriptor: | CALCIUM ION, PP1b, ZINC ION | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2019-02-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Viruses, 12, 2020
|
|
7VJV
 
 | | Human AlkB homolog ALKBH6 in complex with alpha-katoglutarate and Mn | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, MANGANESE (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7VJS
 
 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7VWA
 
 | |
7VW8
 
 | |
7VW9
 
 | |
6JDR
 
 | |
7ENQ
 
 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
7W8S
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
7WA1
 
 | |
