9D88
 
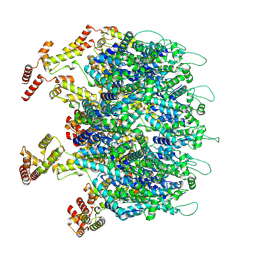 | |
9D6C
 
 | | Gag CA-SP1 immature lattice bound with Lenacapavir and Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
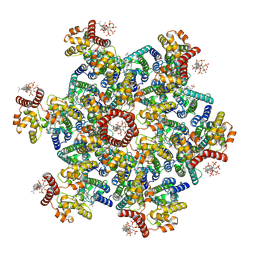
9DWD
 
 | | Gag CA-SP1 immature lattice bound with Lenacapavir from enveloped virus like particles (T8I) | | Descriptor: | Gag, INOSITOL HEXAKISPHOSPHATE, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-10-09 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
8S8C
 
 | | Structure of Kras in complex with inhibitor MK-1084 | | Descriptor: | (5aSa,17aRa)- 20-Chloro-2-[(2S,5R)-2,5-dimethyl-4-(prop-2-enoyl)piperazin-1-yl]-14,17-difluoro-6-(propan-2-yl)-11,12-dihydro-4H-1,18-(ethanediylidene)pyrido[4,3-e]pyrimido[1,6-g][1,4,7,9]benzodioxadiazacyclododecin-4-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Day, P.J, Cleasby, A. | | Deposit date: | 2024-03-06 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of MK-1084: An Orally Bioavailable and Low-Dose KRAS G12C Inhibitor.
J.Med.Chem., 67, 2024
|
|
2ZMN
 
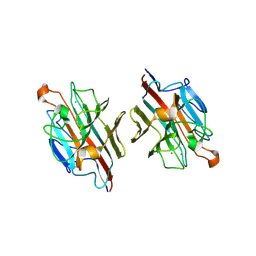 | | Crystal Structure of basic winged bean lectin in complex with Gal-alpha- 1,6 Glc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5D6X
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
2ZMK
 
 | | Crystl structure of Basic Winged bean lectin in complex with Gal-alpha-1,4-Gal-Beta-Ethylene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2ZML
 
 | | Crystal structure of basic winged bean lectin in complex with Gal-ALPHA 1,4 Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3CVE
 
 | | Crystal Structure of the carboxy terminus of Homer1 | | Descriptor: | Homer protein homolog 1 | | Authors: | Hayashi, M.K, Stearns, M.H, Giannini, V, Xu, R.-M, Sala, C, Hayashi, Y. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The postsynaptic density proteins Homer and Shank form a polymeric network structure.
Cell(Cambridge,Mass.), 137, 2009
|
|
6C0S
 
 | |
4UC4
 
 | |
4UD7
 
 | | Structure of the stapled peptide YS-02 bound to MDM2 | | Descriptor: | MDM2, YS-02 | | Authors: | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | Deposit date: | 2014-12-08 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
4UE1
 
 | | Structure of the stapled peptide YS-01 bound to MDM2 | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE MDM2, YS-01 | | Authors: | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | Deposit date: | 2014-12-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
4UP3
 
 | | Crystal structure of the mutant C140S,C286Q thioredoxin reductase from Entamoeba histolytica | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Vieira, D.F. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
5WB6
 
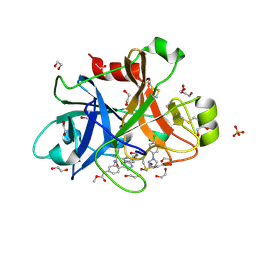 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8W2R
 
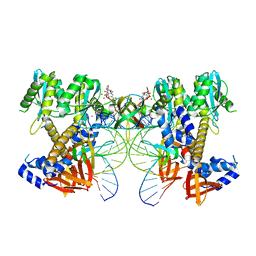 | | HIV-1 P5-IN intasome core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (5'-D(*AP*CP*TP*GP*CP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*CP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*GP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*GP*CP*A)-3'), ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
8W34
 
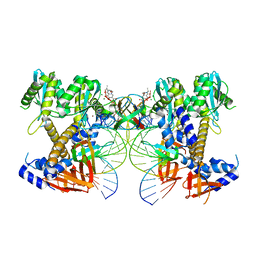 | | HIV-1 intasome core assembled with wild-type integrase, 1F | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (5'-D(*AP*CP*TP*GP*CP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*CP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*GP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*GP*CP*A)-3'), ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
8W09
 
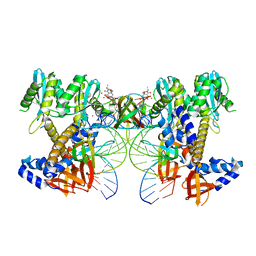 | | HIV-1 wild-type intasome core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, Integrase, MAGNESIUM ION, ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
1ML1
 
 | | PROTEIN ENGINEERING WITH MONOMERIC TRIOSEPHOSPHATE ISOMERASE: THE MODELLING AND STRUCTURE VERIFICATION OF A SEVEN RESIDUE LOOP | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Thanki, N, Zeelen, J.P, Mathieu, M, Jaenicke, R, Abagyan, R.A, Wierenga, R, Schliebs, W. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein engineering with monomeric triosephosphate isomerase (monoTIM): the modelling and structure verification of a seven-residue loop.
Protein Eng., 10, 1997
|
|
7OY5
 
 | | Crystal structure of GSK3Beta in complex with ARN25068 | | Descriptor: | CHLORIDE ION, Glycogen synthase kinase-3 beta, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | Authors: | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Giabbai, B, Storici, P, Ortega, J, Girotto, S, Cavalli, A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
7OY6
 
 | | Crystal structure of human DYRK1A in complex with ARN25068 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | Authors: | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Ortega, J, Girotto, S, Cavalli, A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
4R6R
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
