2O0U
 
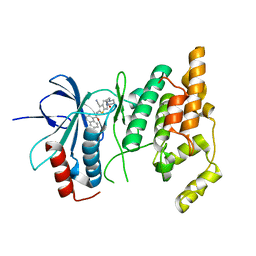 | | Crystal structure of human JNK3 complexed with N-{3-cyano-6-[3-(1-piperidinyl)propanoyl]-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl}-1-naphthalenecarboxamide | | Descriptor: | Mitogen-activated protein kinase 10, N-{3-CYANO-6-[3-(1-PIPERIDINYL)PROPANOYL]-4,5,6,7-TETRAHYDROTHIENO[2,3-C]PYRIDIN-2-YL}1-NAPHTHALENECARBOXAMIDE | | Authors: | Rowland, P, Somers, D. | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N-(3-Cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)amides as potent, selective, inhibitors of JNK2 and JNK3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2O2U
 
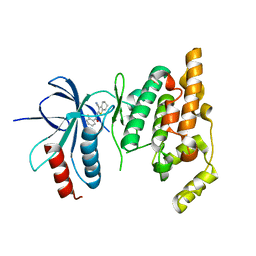 | | Crystal structure of human JNK3 complexed with N-(3-cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)-2-fluorobenzamide | | Descriptor: | Mitogen-activated protein kinase 10, N-(3-cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)-2-fluorobenzamide | | Authors: | Somers, D, Rowland, P. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | N-(3-Cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)amides as potent, selective, inhibitors of JNK2 and JNK3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5X74
 
 | | The crystal Structure PDE delta in complex with compound (R, R)-1g | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-[2-[4-[(2R)-2-(2-fluorophenyl)-4-oxidanylidene-1,2-dihydroquinazolin-3-yl]piperidin-1-yl]ethyl]-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5X73
 
 | | The crystal Structure PDE delta in complex with R-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
6JQH
 
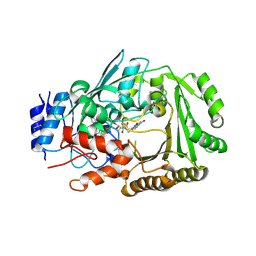 | | Crystal structure of MaDA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MaDA | | Authors: | Du, X.X, Lei, X.G. | | Deposit date: | 2019-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis.
Nat.Chem., 12, 2020
|
|
9GMS
 
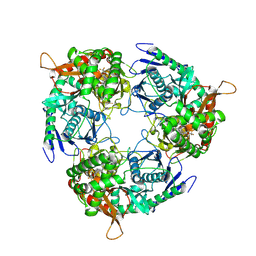 | | Mtb PNPase Rv2783c | | Descriptor: | 1-[[4-[4-[[2-phenyl-5-(trifluoromethyl)-1,3-oxazol-4-yl]carbonylamino]phenyl]phenyl]carbonylamino]cyclopentane-1-carboxylic acid, Polyribonucleotide nucleotidyltransferase, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Griesser, T, Sander, P. | | Deposit date: | 2024-08-29 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Selective inhibition of Mycobacterium tuberculosis GpsI unveils a novel strategy to target the RNA metabolism.
Nucleic Acids Res., 53, 2025
|
|
9GMT
 
 | | Mtb PNPase Rv2783c Mutant L328F | | Descriptor: | 1-[[4-[4-[[2-phenyl-5-(trifluoromethyl)-1,3-oxazol-4-yl]carbonylamino]phenyl]phenyl]carbonylamino]cyclopentane-1-carboxylic acid, Polyribonucleotide nucleotidyltransferase, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Griesser, T, Sander, P. | | Deposit date: | 2024-08-29 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Selective inhibition of Mycobacterium tuberculosis GpsI unveils a novel strategy to target the RNA metabolism.
Nucleic Acids Res., 53, 2025
|
|
5XM8
 
 | |
4HOW
 
 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HK7
 
 | | Crystal structure of Cordyceps militaris IDCase in complex with uracil | | Descriptor: | URACIL, Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4HK5
 
 | | Crystal structure of Cordyceps militaris IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4HOZ
 
 | | The crystal structure of isomaltulose synthase mutant D241A from Erwinia rhapontici NX5 in complex with D-glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HXX
 
 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | Descriptor: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
4HK6
 
 | |
4HPH
 
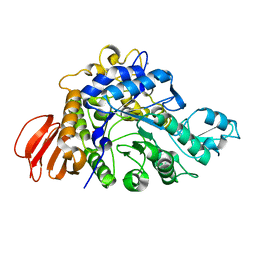 | | The crystal structure of isomaltulose synthase mutant E295Q from Erwinia rhapontici NX5 in complex with its natural substrate sucrose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
5TDK
 
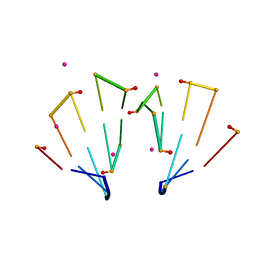 | | RNA decamer duplex with eight 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
5TDJ
 
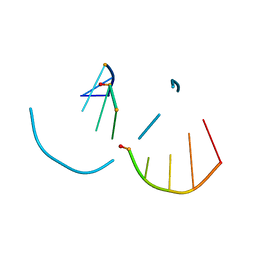 | | RNA decamer duplex with four 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
5U0Q
 
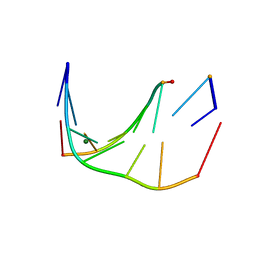 | | RNA-DNA heptamer duplex with one 2'-5'-linkage | | Descriptor: | COBALT (II) ION, DNA/RNA (5'-R(*GP*GP*AP*GP*C)-D(P*T)-R(P*A)-3'), MAGNESIUM ION, ... | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-11-26 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | RNA-DNA heptamer duplex with one 2'-5'-linkage
To Be Published
|
|
5THO
 
 | | Crystal Structure of Mycobacterium Tuberculosis Proteasome in complex with N,C-capped Dipeptide Inhibitor PKS2205 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2016-09-30 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TKO
 
 | | RNA heptamer duplex with one 2'-5'-linkage | | Descriptor: | COBALT (II) ION, RNA (5'-R(*GP*GP*AP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-10-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | RNA heptamer duplex with one 2'-5'-linkage
To Be Published
|
|
4LAL
 
 | | Crystal structure of Cordyceps militaris IDCase D323A mutant in complex with 5-carboxyl-uracil | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, HEXAETHYLENE GLYCOL, Uracil-5-carboxylate decarboxylase, ... | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAM
 
 | | Crystal structure of Cordyceps militaris IDCase D323N mutant in complex with 5-carboxyl-uracil | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, HEXAETHYLENE GLYCOL, Uracil-5-carboxylate decarboxylase, ... | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4HJW
 
 | | Crystal structure of Metarhizium anisopliae IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAN
 
 | | Crystal structure of Cordyceps militaris IDCase H195A mutant | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAO
 
 | | Crystal structure of Cordyceps militaris IDCase H195A mutant (Zn) | | Descriptor: | Cordyceps militaris IDCase, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
