3COJ
 
 | |
2H4O
 
 | | X-ray Crystal Structure of Protein yonK from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR415 | | Descriptor: | YonK protein | | Authors: | Seetharaman, J, Sue, M, Forouhar, F, Ken, C, Bonnie, C, Ma, L, Xiao, R, Acton, T.B, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-24 | | Release date: | 2006-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein from bacillus subtilis (yonk).
To be Published
|
|
3D3U
 
 | | Crystal structure of 4-hydroxybutyrate CoA-transferase (abfT-2) from Porphyromonas gingivalis. Northeast Structural Genomics Consortium target PgR26 | | Descriptor: | 4-hydroxybutyrate CoA-transferase | | Authors: | Forouhar, F, Neely, H, Zhang, X.-Z, Price II, W.N, Hussain, M, Seetharaman, J, Xiao, R, Conover, K, Cunningham, K, Ma, L.-C, Ho, C.K, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of 4-hydroxybutyrate CoA-transferase (abfT-2) from Porphyromonas gingivalis.
To be Published
|
|
3CER
 
 | | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13 | | Descriptor: | Possible exopolyphosphatase-like protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Chen, Y, Neely, H, Seetharaman, J, Shastry, R, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13.
To be Published
|
|
1UYT
 
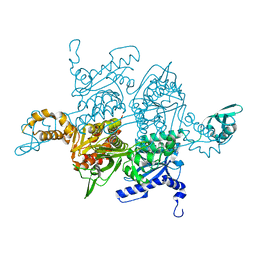 | | Acetyl-CoA carboxylase carboxyltransferase domain | | Descriptor: | ACETYL-COA CARBOXYLASE | | Authors: | Zhang, H, Tweel, B, Tong, L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Inhibition of the Carboxyltransferase Domain of Acetyl-Coenzyme-A Carboxylase by Haloxyfop and Diclofop.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1QR6
 
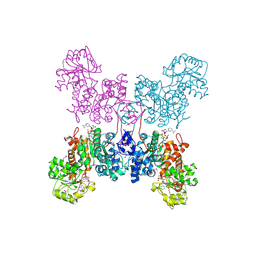 | | HUMAN MITOCHONDRIAL NAD(P)-DEPENDENT MALIC ENZYME | | Descriptor: | MALIC ENZYME 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Xu, Y, Bhargava, G, Wu, H, Loeber, G, Tong, L. | | Deposit date: | 1999-06-18 | | Release date: | 1999-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human mitochondrial NAD(P)(+)-dependent malic enzyme: a new class of oxidative decarboxylases.
Structure, 7, 1999
|
|
1WYZ
 
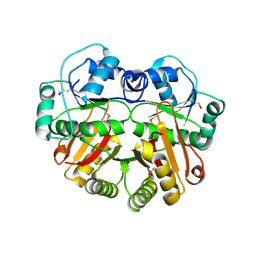 | | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28 | | Descriptor: | putative S-adenosylmethionine-dependent methyltransferase | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S.M, Acton, T, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-21 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28
To be Published
|
|
1SDJ
 
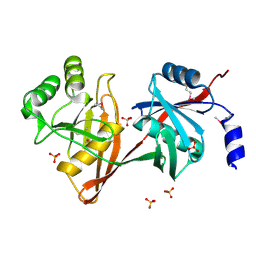 | | X-RAY STRUCTURE OF YDDE_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET25. | | Descriptor: | Hypothetical protein yddE, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Skarina, T, Korniyenko, Y, Savchenko, A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1XBF
 
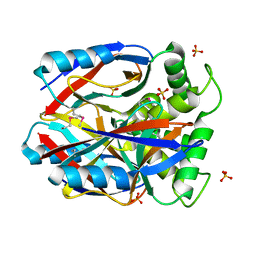 | | X-RAY STRUCTURE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET CAR10 FROM C. ACETOBUTYLICUM | | Descriptor: | Clostridium acetobutylicum Q97KL0, SULFATE ION | | Authors: | Kuzin, A.P, Chen, Y, Vorobiev, S, Yong, W, Acton, T, Ho, C.-K, Conover, K, Cooper, B, Ciano, M, Xiao, R, Montelione, G, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-30 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-RAY STRUCTURE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET CAR10 FROM C. ACETOBUTYLICUM
To be published
|
|
1PJL
 
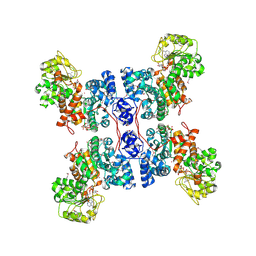 | | Crystal structure of human m-NAD-ME in ternary complex with NAD and Lu3+ | | Descriptor: | LUTETIUM (III) ION, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Yang, Z, Batra, R, Floyd, D.L, Hung, H.-C, Chang, G.-G, Tong, L. | | Deposit date: | 2003-06-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent and competitive inhibition of malic enzymes by lanthanide ions
Biochem.Biophys.Res.Commun., 274, 2000
|
|
1VYT
 
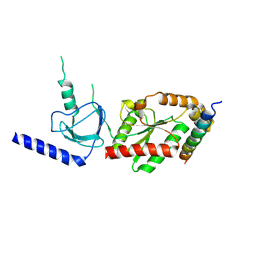 | | beta3 subunit complexed with aid | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1SQS
 
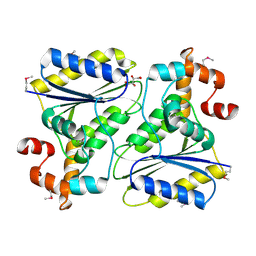 | | X-Ray Crystal Structure Protein SP1951 of Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR27. | | Descriptor: | L(+)-TARTARIC ACID, conserved hypothetical protein | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM, 8, 2007
|
|
1PG6
 
 | | X-Ray Crystal Structure of Protein SPYM3_0169 from Streptococcus pyogenes. Northeast Structural Genomics Consortium Target DR2. | | Descriptor: | CALCIUM ION, Hypothetical protein SpyM3_0169 | | Authors: | Kuzin, A, Lee, I, Edstrom, W, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-05-27 | | Release date: | 2003-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of hypothetical protein SPYM3_0169 from Streptococcus pyogenes
To be Published, 2003
|
|
1SQ4
 
 | | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14 | | Descriptor: | Glyoxylate-induced Protein, THIOCYANATE ION | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14
To be Published
|
|
1XUA
 
 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XSR
 
 | | X-Ray structure of Northeast Structural Genomics Consortium target SfR7 | | Descriptor: | Ureidoglycolate hydrolase | | Authors: | Kuzin, A.P, Vorobiev, S.M, Edstrom, W, Yong, W, Acton, T, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of Northeast
Structural Genomics Consortium target SfR7
To be Published
|
|
1XX6
 
 | | X-ray structure of Clostridium acetobutylicum thymidine kinase with ADP. Northeast Structural Genomics Target CAR26. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thymidine kinase, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Vorobiev, S.M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-11-04 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Clostridium acetobutylicum thymidine kinase with ADP. Northeast Structural Genomics Target CAR26.
To be Published
|
|
1SPV
 
 | | Crystal Structure of the Putative Phosphatase of Escherichia coli, Northeast Structural Genomoics Target ER58 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, putative polyprotein/phosphatase | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Phosphatase of Escherichia coli, Northeast Structural Genomoics Target ER58
To be Published
|
|
1SQ1
 
 | | Crystal Structure of the Chorismate Synthase from Campylobacter jejuni, Northeast Structural Genomics Target BR19 | | Descriptor: | Chorismate synthase, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Chorismate Synthase from Campylobacter jejuni, Northeast Structural Genomics Target BR19
To be Published
|
|
1SQH
 
 | | X-RAY STRUCTURE OF DROSOPHILA MALONOGASTER PROTEIN Q9VR51 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET FR87. | | Descriptor: | hypothetical protein CG14615-PA | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Acton, T.B, Xiao, R, Cooper, B, Ho, C.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-18 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-RAY STRUCTURE OF DROSOPHILA MALONOGASTER PROTEIN Q9VR51 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET FR87
To be Published
|
|
1XKN
 
 | | Crystal Structure of the Putative Peptidyl-arginine Deiminase from Chlorobium tepidum, NESG Target CtR21 | | Descriptor: | CHLORIDE ION, SODIUM ION, putative peptidyl-arginine deiminase | | Authors: | Forouhar, F, Befeler, A.R, Vorobiev, S.M, Hussain, M, Ciano, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Putative Peptidyl-arginine Deiminase from Chlorobium tepidum, NESG Target CtR21
To be Published
|
|
1XUV
 
 | | X-Ray Crystal Structure of Protein MM0500 from Methanosarcina mazei. Northeast Structural Genomics Consortium Target MaR10. | | Descriptor: | hypothetical protein MM0500 | | Authors: | Forouhar, F, Abashidze, M, Ciano, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Methanosarcina mazei, Northeast Strcutural Genomics Target MaR10
To be Published
|
|
1XMD
 
 | | M335V mutant structure of mouse carnitine octanoyltransferase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peroxisomal carnitine O-octanoyltransferase | | Authors: | Jogl, G, Hsiao, Y.S, Tong, L. | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity.
J.Biol.Chem., 280, 2005
|
|
1XMC
 
 | | C323M mutant structure of mouse carnitine octanoyltransferase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peroxisomal carnitine O-octanoyltransferase | | Authors: | Jogl, G, Hsiao, Y.S, Tong, L. | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity.
J.Biol.Chem., 280, 2005
|
|
1XL8
 
 | | Crystal structure of mouse carnitine octanoyltransferase in complex with octanoylcarnitine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARNITINE, OCTANOYLCARNITINE, ... | | Authors: | Jogl, G, Hsiao, Y.S, Tong, L. | | Deposit date: | 2004-09-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity.
J.Biol.Chem., 280, 2005
|
|
