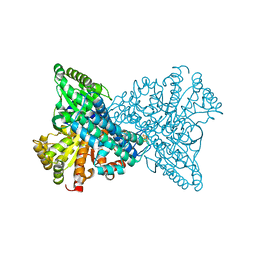
7XDP
 
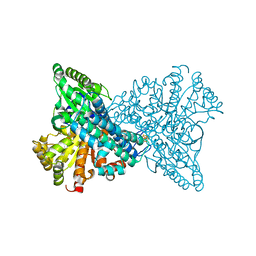 | |
7XDM
 
 | | ChCODH2 A559W mutant in anaerobic condition | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Heo, Y.Y, Kim, S.M. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | O2-tolerant CO dehydrogenase via tunnel redesign for the removal of CO from industrial flue gas
Nat Catal, 5, 2022
|
|
7XDN
 
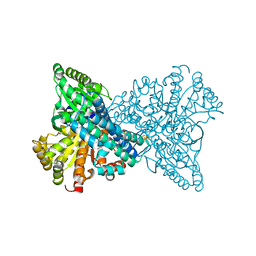 | |
3JQD
 
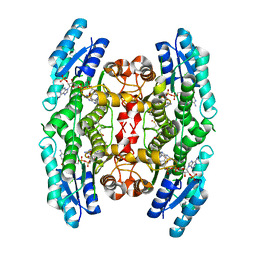 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-4-oxo-6-phenyl-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile (DX7) | | Descriptor: | 2-amino-4-oxo-6-phenyl-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQA
 
 | |
3JQC
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-6-bromo-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile (JU2) | | Descriptor: | 2-amino-6-bromo-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQ9
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-6-(1,3-benzodioxol-5-yl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile (AX1) | | Descriptor: | 2-amino-6-(1,3-benzodioxol-5-yl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQG
 
 | |
3JQ8
 
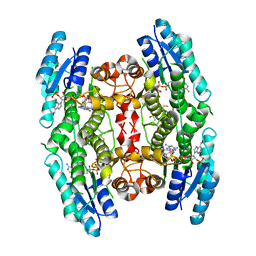 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 6,7,7-trimethyl-7,8-dihydropteridine-2,4-diamine (DX3) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6,7,7-trimethyl-7,8-dihydropteridine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQB
 
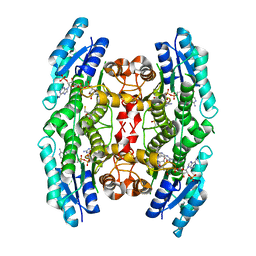 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-5-(2-phenylethyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one (DX6) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-amino-5-(2-phenylethyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQ6
 
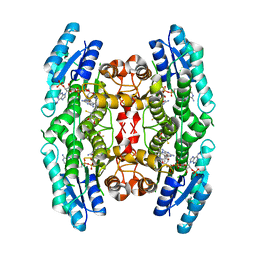 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 6,7-bis(1-methylethyl)pteridine-2,4-diamine (DX1) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6,7-bis(1-methylethyl)pteridine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQE
 
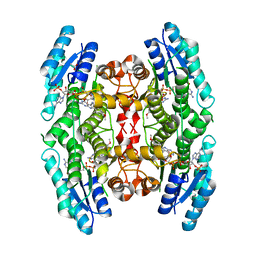 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-6-(4-methoxyphenyl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile (DX8) | | Descriptor: | 2-amino-6-(4-methoxyphenyl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQ7
 
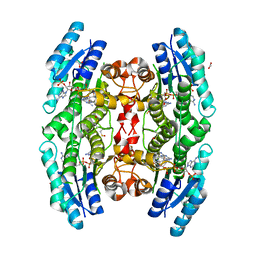 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 6-phenylpteridine-2,4,7-triamine (DX2) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-phenylpteridine-2,4,7-triamine, ACETATE ION, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3JQF
 
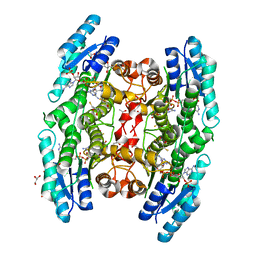 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 1,3,5-triazine-2,4,6-triamine (AX2) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1,3,5-triazine-2,4,6-triamine, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3LDJ
 
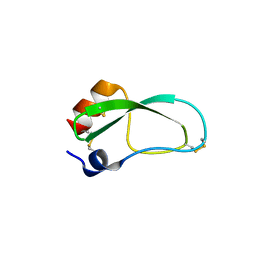 | | Crystal structure of aprotinin in complex with sucrose octasulfate: unusual interactions and implication for heparin binding | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACETATE ION, Pancreatic trypsin inhibitor | | Authors: | Yang, I.S, Kim, T.G, Park, B.S, Kim, K.H. | | Deposit date: | 2010-01-13 | | Release date: | 2010-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding.
Biochem.Biophys.Res.Commun., 2010
|
|
3LDM
 
 | |
6CGW
 
 | |
6CHC
 
 | | JzTx-V toxin peptide, wild-type | | Descriptor: | Beta/kappa-theraphotoxin-Cg2a | | Authors: | Jordan, J.B. | | Deposit date: | 2018-02-22 | | Release date: | 2018-05-16 | | Method: | SOLUTION NMR | | Cite: | Pharmacological characterization of potent and selective NaV1.7 inhibitors engineered from Chilobrachys jingzhao tarantula venom peptide JzTx-V.
PLoS ONE, 13, 2018
|
|
5IT2
 
 | | Structure of a transglutaminase 2-specific autoantibody 693-10-B06 Fab fragment | | Descriptor: | heavy chain, light chain | | Authors: | Chen, X, Dalhus, B, Hnida, K, Iversen, R, Sollid, L.M. | | Deposit date: | 2016-03-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions
Nat. Med., 18, 2012
|
|
5CEZ
 
 | |
5K52
 
 | |
5CEY
 
 | |
5CEX
 
 | |
5K53
 
 | |
5D6Y
 
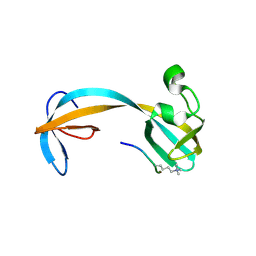 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
