3M9A
 
 | | Protein structure of type III plasmid segregation TubR | | Descriptor: | Putative DNA-binding protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3O52
 
 | | Structure of the E.coli GDP-mannose hydrolase (yffh) in complex with tartrate | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, GDP-mannose pyrophosphatase nudK, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3DNU
 
 | | structure of MDT protein | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Protein hipA | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3O6Z
 
 | | Structure of the D152A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3FBR
 
 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3O69
 
 | | Structure of the E100A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GDP-mannose pyrophosphatase nudK, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3O61
 
 | | Structure of the E100A E.coli GDP-mannose hydrolase (yffh) in complex with GDP-mannose and Mg++ | | Descriptor: | CHLORIDE ION, GDP-mannose pyrophosphatase nudK, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | Descriptor: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
7W6L
 
 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6A
 
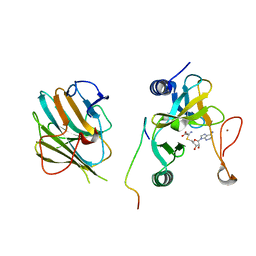 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
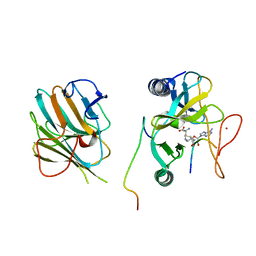 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
3TPT
 
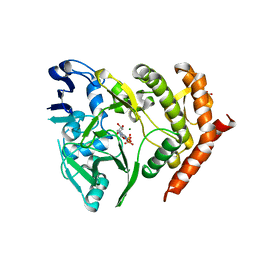 | | Structure of HipA(D309Q) bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPB
 
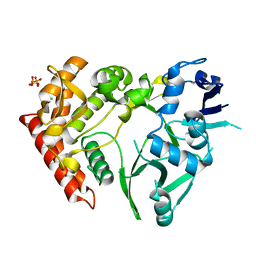 | | Structure of HipA(S150A) | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | Authors: | schumacher, M.A. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
2P3U
 
 | |
3TPE
 
 | | The phipa p3121 structure | | Descriptor: | Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
2GL7
 
 | | Crystal Structure of a beta-catenin/BCL9/Tcf4 complex | | Descriptor: | B-cell lymphoma 9 protein, Beta-catenin, Transcription factor 7-like 2 | | Authors: | Sampietro, J. | | Deposit date: | 2006-04-04 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a beta-Catenin/BCL9/Tcf4 Complex.
Mol.Cell, 24, 2006
|
|
3TPD
 
 | | Structure of pHipA, monoclinic form | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPV
 
 | | Structure of pHipA bound to ADP | | Descriptor: | ADENINE, SULFATE ION, Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T.M, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
2P3T
 
 | |
7Y7J
 
 | | SARS-CoV-2 S trimer in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7Y7K
 
 | | SARS-CoV-2 RBD in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, Spike protein S1 | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
3Q4I
 
 | | Crystal structure of CDP-Chase in complex with Gd3+ | | Descriptor: | GADOLINIUM ION, Phosphohydrolase (MutT/nudix family protein) | | Authors: | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-12-23 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
