3BZ6
 
 | |
3BYW
 
 | | Crystal structure of an extracellular domain of arabinofuranosyltransferase from Corynebacterium diphtheriae | | Descriptor: | ACETATE ION, Putative arabinofuranosyltransferase, ZINC ION | | Authors: | Tan, K, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of an extracellular domain of arabinofuranosyltransferase from Corynebacterium diphtheriae.
To be Published
|
|
3CCY
 
 | |
3CDL
 
 | |
3CQY
 
 | | Crystal structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1 | | Descriptor: | Anhydro-N-acetylmuramic acid kinase, CHLORIDE ION, SUCCINIC ACID | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1.
To be Published
|
|
3D3Y
 
 | | Crystal structure of a conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Uncharacterized protein | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-13 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of a conserved protein from Enterococcus faecalis V583.
To be Published
|
|
4NZP
 
 | | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Argininosuccinate synthase | | Authors: | Tan, K, Gu, M, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4NOC
 
 | |
3CJ8
 
 | | Crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Enterococcus faecalis V583 | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-acetyltransferase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Enterococcus faecalis V583.
To be Published
|
|
4M7O
 
 | |
3COO
 
 | |
3D7L
 
 | |
3D6K
 
 | | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Tan, K, Zhang, R, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae
To be Published
|
|
1GEH
 
 | | CRYSTAL STRUCTURE OF ARCHAEAL RUBISCO (RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE) | | Descriptor: | RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE, SULFATE ION | | Authors: | Kitano, K, Maeda, N, Fukui, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2000-11-13 | | Release date: | 2001-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Novel-Type Archaeal Rubisco with Pentagonal Symmetry
Structure, 9, 2001
|
|
1GP9
 
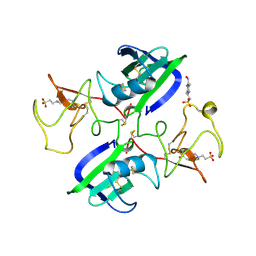 | | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Watanabe, K, Chirgadze, D.Y, Lietha, D, Gherardi, E, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A New Crystal Form of the Nk1 Splice Variant of Hgf/Sf Demonstrates Extensive Hinge Movement and Suggests that the Nk1 Dimer Originates by Domain Swapping
J.Mol.Biol., 319, 2002
|
|
4XXI
 
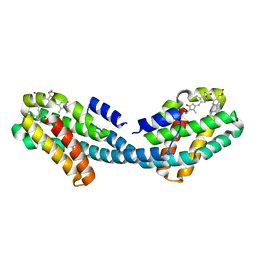 | | Crystal structure of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | Descriptor: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | Authors: | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | Deposit date: | 2015-01-30 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1WE0
 
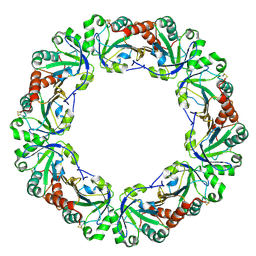 | | Crystal structure of peroxiredoxin (AhpC) from Amphibacillus xylanus | | Descriptor: | AMMONIUM ION, alkyl hydroperoxide reductase C | | Authors: | Kitano, K, Kita, A, Hakoshima, T, Niimura, Y, Miki, K. | | Deposit date: | 2004-05-21 | | Release date: | 2005-03-29 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of decameric peroxiredoxin (AhpC) from Amphibacillus xylanus
Proteins, 59, 2005
|
|
4XXK
 
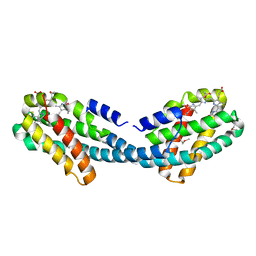 | | Crystal structure of the Semet-derivative of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | Descriptor: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | Authors: | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | Deposit date: | 2015-01-30 | | Release date: | 2015-12-16 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6F9L
 
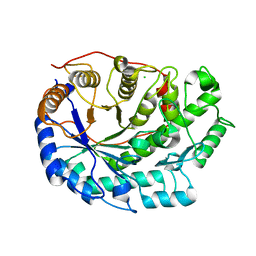 | | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose | | Descriptor: | Beta-amylase, CHLORIDE ION, alpha-D-glucopyranose-(1-4)-3-deoxy-3-fluoro-alpha-D-glucopyranose | | Authors: | Tantanarat, K, Stevenson, C.E.M, Rejzek, M, Lawson, D.M, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose
To be published
|
|
1NFT
 
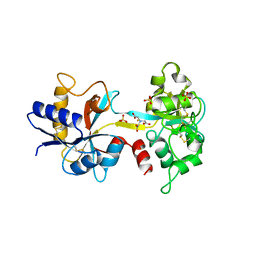 | | OVOTRANSFERRIN, N-TERMINAL LOBE, IRON LOADED OPEN FORM | | Descriptor: | FE (III) ION, NITRILOTRIACETIC ACID, PROTEIN (OVOTRANSFERRIN), ... | | Authors: | Mizutani, K, Yamashita, H, Kurokawa, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
1IYY
 
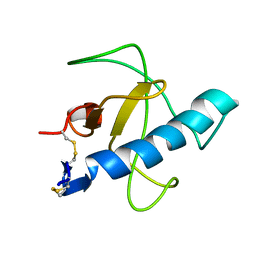 | | NMR STRUCTURE OF Gln25-RIBONUCLEASE T1, 24 STRUCTURES | | Descriptor: | RIBONUCLEASE T1 | | Authors: | Hatano, K, Kojima, M, Suzuki, E, Tanokura, M, Takahashi, K. | | Deposit date: | 2002-09-12 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Determination of the NMR structure of Gln25-ribonuclease T1.
Biol. Chem., 384, 2003
|
|
7VRJ
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF Allochromatium tepidum | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Tani, K, Kobayashi, K, Hosogi, N, Ji, X.-C, Nagashima, S, Nagashima, K.V.P, Tsukatani, Y, Kanno, R, Hall, M, Yu, L.-J, Ishikawa, I, Okura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-10-23 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | A Ca 2+ -binding motif underlies the unusual properties of certain photosynthetic bacterial core light-harvesting complexes.
J.Biol.Chem., 298, 2022
|
|
7E3Y
 
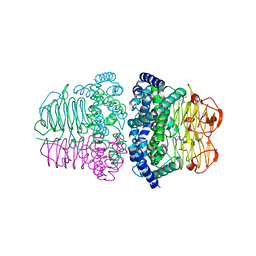 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with cysteine | | Descriptor: | CYSTEINE, Serine acetyltransferase | | Authors: | Momitani, K, Shiba, T, Sawa, T, Ono, K, Hurukawa, S. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of serine acetyltransferase from Salmonella typhimurium
To Be Published
|
|
1X26
 
 | | Solution structure of the AA-mismatch DNA complexed with naphthyridine-azaquinolone | | Descriptor: | 5'-D(*CP*AP*TP*TP*CP*AP*GP*TP*TP*AP*G)-3', 5'-D(*CP*TP*AP*AP*CP*AP*GP*AP*AP*TP*G)-3', N~3~-{3-[(7-METHYL-1,8-NAPHTHYRIDIN-2-YL)AMINO]-3-OXOPROPYL}-N~1~-[(7-OXO-7,8-DIHYDRO-1,8-NAPHTHYRIDIN-2-YL)METHYL]-BET A-ALANINAMIDE | | Authors: | Nakatani, K, Hagihara, S, Goto, Y, Kobori, A, Hagihara, M, Hayashi, G, Kyo, M, Nomura, M, Mishima, M, Kojima, C. | | Deposit date: | 2005-04-20 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats
Nat.Chem.Biol., 1, 2005
|
|
7XXF
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodopila globiformis | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E},28~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26,28-dodecaen-5-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Tani, K, Kanno, R, Kurosawa, K, Takaichi, S, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-05-30 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins.
Commun Biol, 5, 2022
|
|
