3KEE
 
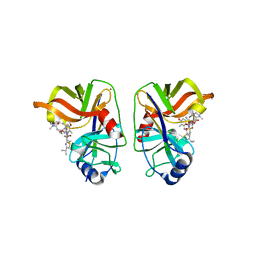 | | HCV NS3/NS4A complexed with Non-covalent macrocyclic compound TMC435 | | Descriptor: | (2R,3aR,10Z,11aS,12aR,14aR)-N-(cyclopropylsulfonyl)-2-({7-methoxy-8-methyl-2-[4-(1-methylethyl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-4,14-dioxo-2,3,3a,4,5,6,7,8,9,11a,12,13,14,14a-tetradecahydrocyclopenta[c]cyclopropa[g][1,6]diazacyclotetradecine-12a(1H)-carboxamide, 19-mer peptide from Genome polyprotein, GLYCEROL, ... | | Authors: | Lindberg, J.D, Nystrom, S, Cummings, M.D. | | Deposit date: | 2009-10-26 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Induced-Fit Binding of the Macrocyclic Noncovalent Inhibitor TMC435 to its HCV NS3/NS4A Protease Target
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
8OZ3
 
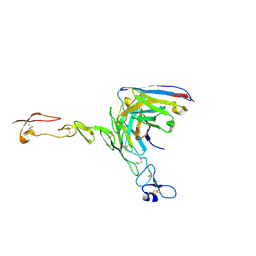 | | Crystal structure of scFv ATOR 1017 bound to human 4-1BB | | Descriptor: | Single chain Fv, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Hakansson, M, Von Schantz, L, Rose, N. | | Deposit date: | 2023-05-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | ATOR-1017 (evunzekibart), an Fc-gamma receptor conditional 4-1BB agonist designed for optimal safety and efficacy, activates exhausted T cells in combination with anti-PD-1.
Cancer Immunol.Immunother., 72, 2023
|
|
3KTR
 
 | |
1J6X
 
 | | CRYSTAL STRUCTURE OF HELICOBACTER PYLORI LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
3KYR
 
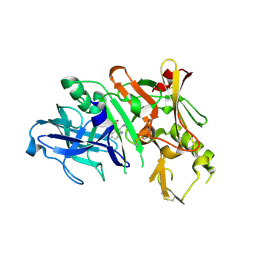 | | Bace-1 in complex with a norstatine type inhibitor | | Descriptor: | 3-[[(2S)-2-[[[(2S)-2-[[(2S)-2-[[(2S)-2-azanyl-3-(1H-1,2,3,4-tetrazol-5-ylcarbonylamino)propanoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-pentanoyl]amino]methyl]-2-hydroxy-4-phenyl-butanoyl]amino]benzoic acid, Beta-secretase 1 | | Authors: | Lindberg, J.D, Borkakoti, N, Derbyshire, D, Nystrom, S. | | Deposit date: | 2009-12-07 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of a-phenylnorstatine and a-benzylnorstatine as transition state isostere motifs in the search for new BACE-1 inhibiotrs
To be Published
|
|
3KTP
 
 | | Structural basis of GW182 recognition by poly(A)-binding protein | | Descriptor: | Polyadenylate-binding protein 1, Trinucleotide repeat-containing gene 6C protein | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2009-11-25 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein.
J.Biol.Chem., 285, 2010
|
|
3KUS
 
 | |
3KUR
 
 | |
3SQO
 
 | | PCSK9 J16 Fab complex | | Descriptor: | J16 Heavy chain, J16 Light chain, PCSK9 prodomain, ... | | Authors: | Strop, P. | | Deposit date: | 2011-07-05 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Proprotein convertase substilisin/kexin type 9 antagonism reduces low-density lipoprotein cholesterol in statin-treated hypercholesterolemic nonhuman primates.
J.Pharmacol.Exp.Ther., 340, 2012
|
|
1J6W
 
 | | CRYSTAL STRUCTURE OF HAEMOPHILUS INFLUENZAE LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
3SMJ
 
 | | Human poly(ADP-ribose) polymerase 14 (Parp14/Artd8) - catalytic domain in complex with a pyrimidine-like inhibitor | | Descriptor: | 2-methyl-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one, GLYCEROL, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
3V2B
 
 | | Human poly(adp-ribose) polymerase 15 (ARTD7, BAL3), macro domain 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 15, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Kotzcsh, A, Kraulis, P, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van den berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
6HM0
 
 | | Crystal structure of human BRD9 bromodomain in complex with a PROTAC | | Descriptor: | (2~{S},4~{S})-1-[(2~{S})-2-[2-[2-[2-[4-[[2,6-dimethoxy-4-(2-methyl-1-oxidanylidene-2,7-naphthyridin-4-yl)phenyl]methyl]piperazin-1-yl]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 9 | | Authors: | Hughes, S.J, Zoppi, V, Ciulli, A. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Iterative Design and Optimization of Initially Inactive Proteolysis Targeting Chimeras (PROTACs) Identify VZ185 as a Potent, Fast, and Selective von Hippel-Lindau (VHL) Based Dual Degrader Probe of BRD9 and BRD7.
J.Med.Chem., 62, 2019
|
|
4NYT
 
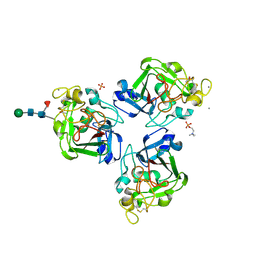 | | L-Ficolin Complexed to Phosphocholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Laffly, E, Gaboriaud, C, Martin, L, Thielens, N. | | Deposit date: | 2013-12-11 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human L-ficolin recognizes phosphocholine moieties of pneumococcal teichoic Acid
J.Immunol., 193, 2014
|
|
1VGT
 
 | |
1VH5
 
 | |
1VHE
 
 | |
1VHS
 
 | |
1VI0
 
 | | Crystal structure of a transcriptional regulator | | Descriptor: | DODECYL-COA, transcriptional regulator | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHC
 
 | |
1VI8
 
 | |
1VH1
 
 | | Crystal structure of CMP-KDO synthetase | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHM
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Protein yebR | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHX
 
 | |
1VI6
 
 | | Crystal structure of ribosomal protein S2P | | Descriptor: | 30S ribosomal protein S2P, SODIUM ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
