1N9Y
 
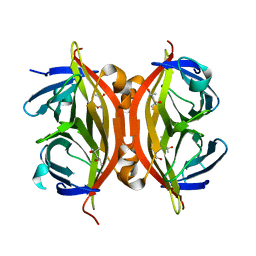 | | Streptavidin Mutant S27A at 1.5A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-11-26 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1RH8
 
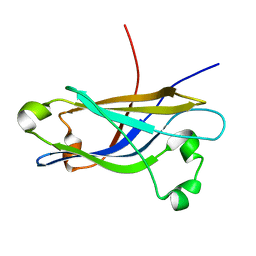 | | Three-dimensional structure of the calcium-free Piccolo C2A-domain | | Descriptor: | Piccolo protein | | Authors: | Garcia, J, Gerber, S.H, Sugita, S, Sudhof, T.C, Rizo, J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the Piccolo C2A domain regulated by alternative splicing.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1NBX
 
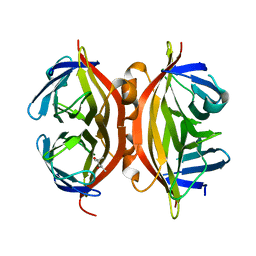 | | Streptavidin Mutant Y43A at 1.70A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2YZA
 
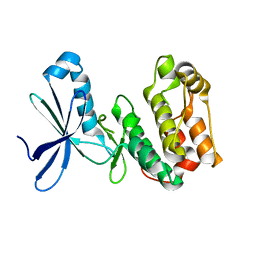 | | Crystal structure of kinase domain of Human 5'-AMP-activated protein kinase alpha-2 subunit mutant (T172D) | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Saijo, S, Takagi, T, Yoshikawa, S, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for compound C inhibition of the human AMP-activated protein kinase alpha 2 subunit kinase domain
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2Z0A
 
 | | Crystal structure of RNA-binding domain of NS1 from influenza A virus A/crow/Kyoto/T1/2004(H5N1) | | Descriptor: | GLYCINE, Nonstructural protein 1, SUCCINIC ACID | | Authors: | Saijo, S, Kishishita, S, Kamo-Uchikubo, T, Terada, T, Shirouzu, M, Ito, H, Ito, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of RNA-binding domain of NS1 from influenza A virus A/crow/Kyoto/T1/2004(H5N1)
To be Published
|
|
2YYH
 
 | |
2YZ8
 
 | | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556) | | Descriptor: | Obscurin | | Authors: | Saijo, S, Ohsawa, N, Nishino, A, Kishishita, S, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556)
To be Published
|
|
2Z0B
 
 | | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434) | | Descriptor: | PHOSPHATE ION, Putative glycerophosphodiester phosphodiesterase 5 | | Authors: | Saijo, S, Nishino, A, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434)
To be Published
|
|
2Z14
 
 | | Crystal structure of the N-terminal DUF1126 in human ef-hand domain containing 2 protein | | Descriptor: | EF-hand domain-containing family member C2 | | Authors: | Saito, K, Olsen, S, Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of the N-terminal DUF1126 in human ef-hand domain containing 2 protein
To be Published
|
|
2Z16
 
 | | Crystal structure of Matrix protein 1 from influenza A virus A/crow/Kyoto/T1/2004(H5N1) | | Descriptor: | Matrix protein 1 | | Authors: | Saijo, S, Kishishita, S, Uchikubo-Kamo, T, Terada, T, Shirouzu, M, Ito, H, Ito, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of Matrix protein 1 from influenza A virus A/crow/Kyoto/T1/2004(H5N1)
To be Published
|
|
1ZB1
 
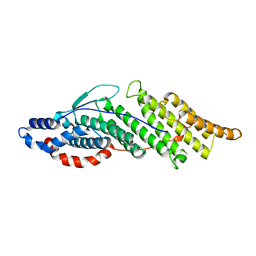 | | Structure basis for endosomal targeting by the Bro1 domain | | Descriptor: | BRO1 protein | | Authors: | Kim, J, Sitaraman, S, Hierro, A, Beach, B.M, Odorizzi, G, Hurley, J.H. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for endosomal targeting by the Bro1 domain.
Dev.Cell, 8, 2005
|
|
2Z1A
 
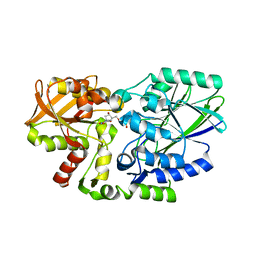 | | Crystal structure of 5'-nucleotidase precursor from Thermus thermophilus HB8 | | Descriptor: | 5'-nucleotidase, PHOSPHATE ION, THYMIDINE, ... | | Authors: | Nakagawa, N, Kishishita, S, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of 5'-nucleotidase precursor from Thermus thermophilus HB8
To be Published
|
|
2YV6
 
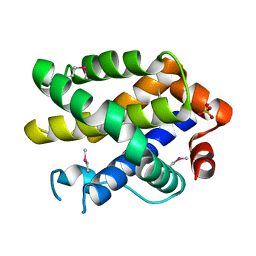 | | Crystal structure of human Bcl-2 family protein Bak | | Descriptor: | Bcl-2 homologous antagonist/killer, SULFATE ION | | Authors: | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator.
J.Struct.Biol., 166, 2009
|
|
1HR9
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Malate Dehydrogenase Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALATE DEHYDROGENASE, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
3VGK
 
 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus | | Descriptor: | Glucokinase, SULFATE ION, ZINC ION | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
7XBD
 
 | | Cryo-EM structure of human galanin receptor 2 | | Descriptor: | Galanin, Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ishimoto, N, Kita, S, Park, S.Y. | | Deposit date: | 2022-03-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of the human galanin receptor 2 bound to galanin and Gq reveals the basis of ligand specificity and how binding affects the G-protein interface.
Plos Biol., 20, 2022
|
|
1IVV
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Early intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVU
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Initial intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1HR7
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant | | Descriptor: | MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ZINC ION | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
3VGM
 
 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose | | Descriptor: | Glucokinase, POTASSIUM ION, ZINC ION, ... | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
3VGL
 
 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose and AMPPNP | | Descriptor: | Glucokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
1HR6
 
 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1IVW
 
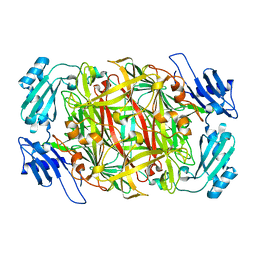 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Late intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVX
 
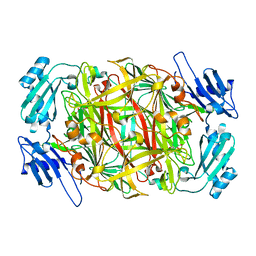 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Holo form generated by biogenesis in crystal. | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
