9LNQ
 
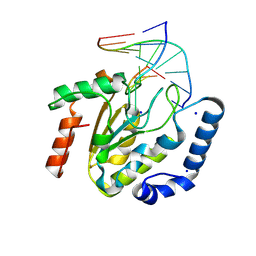 | | The hUNG bound to DNA product embedding 4primer-OCH3-dU | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(KBC)P*AP*TP*CP*TP*T)-3'), SODIUM ION, ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
9QFU
 
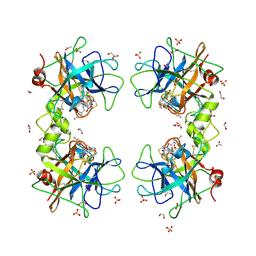 | | Human Tryptase beta-2 (hTPSB2) complexed with covalent inhibitor Compound #1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porta, A, Manelfi, C, Talarico, C, Beccari, A.R, Brindisi, M, Summa, V, Iaconis, D, Gobbi, M, Beeg, M. | | Deposit date: | 2025-03-12 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Integrating Surface Plasmon Resonance and Docking Analysis for Mechanistic Insights of Tryptase Inhibitors.
Molecules, 30, 2025
|
|
2VLW
 
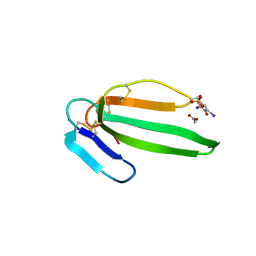 | | Crystal structure of the muscarinic toxin MT7 diiodoTYR51 derivative. | | Descriptor: | ACETATE ION, MUSCARINIC M1-TOXIN1, SULFATE ION | | Authors: | Menez, R, Granata, V, Mourier, G, Fruchart-Gaillard, C, Menez, A, Servant, D, Stura, E.A. | | Deposit date: | 2008-01-17 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Different Interactions between Mt7 Toxin and the Human Muscarinic M1 Receptor in its Free and N-Methylscopolamine-Occupied States.
Mol.Pharmacol., 74, 2008
|
|
2VI4
 
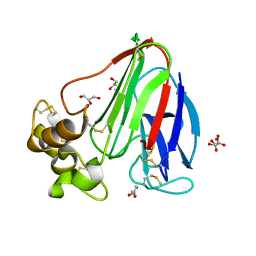 | |
2V5W
 
 | | Crystal structure of HDAC8-substrate complex | | Descriptor: | GLYCYL-GLYCYL-GLYCINE, HISTONE DEACETYLASE 8, PEPTIDIC SUBSTRATE, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
9QFV
 
 | | Human Tryptase beta-2 (hTPSB2) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Porta, A, Manelfi, C, Talarico, C, Beccari, A.R, Brindisi, M, Summa, V, Iaconis, D, Gobbi, M, Beeg, M. | | Deposit date: | 2025-03-12 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Integrating Surface Plasmon Resonance and Docking Analysis for Mechanistic Insights of Tryptase Inhibitors.
Molecules, 30, 2025
|
|
2VHK
 
 | |
8RWZ
 
 | | Open non-crosslinked structure Brd4BD2-MZ1-(NEDD8)-CRL2VHL | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Cullin-2, ... | | Authors: | Crowe, C, Nakasone, M.A, Ciulli, A. | | Deposit date: | 2024-02-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of degrader-targeted protein ubiquitinability.
Sci Adv, 10, 2024
|
|
8RX0
 
 | | (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Crowe, C, Nakasone, M.A, Ciulli, A. | | Deposit date: | 2024-02-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of degrader-targeted protein ubiquitinability.
Sci Adv, 10, 2024
|
|
6TK2
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 1ms structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TQL
 
 | | Cryo-EM of elastase-treated human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
8RPQ
 
 | |
8RQ6
 
 | |
6TK1
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 20ms structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
4UDD
 
 | | GR in complex with desisobutyrylciclesonide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DESISOBUYTYRYL CICLESONIDE, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDC
 
 | | GR in complex with dexamethasone | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DEXAMETHASONE, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
6TK5
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 800fs+2ps structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
8RO0
 
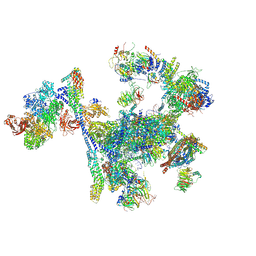 | | Structure of the C. elegans Intron Lariat Spliceosome primed for disassembly (ILS') | | Descriptor: | Cell division cycle 5-like protein, Coiled-coil domain-containing protein 12, GCF C-terminal domain-containing protein, ... | | Authors: | Vorlaender, M.K, Rothe, P, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
4UDA
 
 | | MR in complex with dexamethasone | | Descriptor: | DEXAMETHASONE, GLYCEROL, MINERALOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
6TK7
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: Dark structure in acidic conditions | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
8RJJ
 
 | | HCV E1/E2 homodimer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, ... | | Authors: | Augestad, E.H, Olesen, C.H, Groenberg, C, Soerensen, A, Velazquez-Moctezuma, R, Fanalista, M, Bukh, J, Wang, K, Gourdon, P, Prentoe, J. | | Deposit date: | 2023-12-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 633, 2024
|
|
8RK0
 
 | | HCV E1/E2 homodimer complex, ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HCV E1, ... | | Authors: | Augestad, E.H, Olesen, C.H, Groenberg, C, Soerensen, A, Velazquez-Moctezuma, R, Fanalista, M, Bukh, J, Wang, K, Gourdon, P, Prentoe, J. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 633, 2024
|
|
6TQK
 
 | | Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
1KK8
 
 | | SCALLOP MYOSIN (S1-ADP-BeFx) IN THE ACTIN-DETACHED CONFORMATION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Himmel, M, Gourinath, S, Reshetnikova, L, Shen, Y, Szent-Gyorgyi, G, Cohen, C. | | Deposit date: | 2001-12-06 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KQX
 
 | | Crystal structure of apo-CRBP from zebrafish | | Descriptor: | Cellular retinol-binding protein | | Authors: | Calderone, V, Folli, C, Marchesani, A, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and structural analysis of a zebrafish apo and holo cellular retinol-binding protein.
J.Mol.Biol., 321, 2002
|
|
