2F84
 
 | |
2FJM
 
 | | The structure of phosphotyrosine phosphatase 1B in complex with compound 2 | | Descriptor: | (4-{(2S,4E)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-2-[4-(METHOXYCARBONYL)PHENYL]-5-PHENYLPENT-4-ENYL}PHENYL)(DIFLUORO)METHYLPHOSPHONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Asante-Appiah, E, Patel, S, Desponts, C, Taylor, J.M, Lau, C, Dufresne, C, Therien, M, Friesen, R, Becker, J.W, Leblanc, Y, Scapin, G. | | Deposit date: | 2006-01-03 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformation-assisted inhibition of protein-tyrosine phosphatase-1B elicits inhibitor selectivity over T-cell protein-tyrosine phosphatase.
J.Biol.Chem., 281, 2006
|
|
2HH9
 
 | | Thiamin pyrophosphokinase from Candida albicans | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, MAGNESIUM ION, Thiamin pyrophosphokinase | | Authors: | Abergel, C, Santini, S, Monchois, V, Rousselle, T, Claverie, J.M, Bacterial targets at IGS-CNRS, France (BIGS) | | Deposit date: | 2006-06-28 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of CA1462, the Candida albicans thiamine pyrophosphokinase.
Bmc Struct.Biol., 8, 2008
|
|
2HJJ
 
 | | Solution NMR structure of protein ykfF from Escherichia coli. Northeast Structural Genomics target ER397. | | Descriptor: | Hypothetical protein ykfF | | Authors: | Swapna, G.V.T, Aramini, J.M, Zhao, L, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Liu, J, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein ykfF from Escherichia coli. Northeast Structural Genomics target ER397.
To be Published
|
|
2HIV
 
 | | ATP-dependent DNA ligase from S. solfataricus | | Descriptor: | Thermostable DNA ligase | | Authors: | Pascal, J.M, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2HIX
 
 | |
2E5L
 
 | | A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine- Dalgarno interaction | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kaminishi, T, Wilson, D.N, Takemoto, C, Harms, J.M, Kawazoe, M, Schluenzen, F, Hanawa-Suetsugu, K, Shirouzu, M, Fucini, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-21 | | Release date: | 2007-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction
Structure, 15, 2007
|
|
2EG7
 
 | | The crystal structure of E. coli dihydroorotase complexed with HDDP | | Descriptor: | 2-OXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4,6-DICARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design
J.Mol.Biol., 370, 2007
|
|
2EG6
 
 | |
2HU9
 
 | | X-ray structure of the Archaeoglobus fulgidus CopZ N-terminal Domain | | Descriptor: | ACETIC ACID, FE2/S2 (INORGANIC) CLUSTER, Mercuric transport protein periplasmic component, ... | | Authors: | Sazinsky, M.H, LeMoine, B, Arguello, J.M, Rosenzweig, A.C. | | Deposit date: | 2006-07-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Characterization and structure of a Zn2+ and [2Fe-2S]-containing copper chaperone from Archaeoglobus fulgidus.
J.Biol.Chem., 282, 2007
|
|
2HT1
 
 | |
2IA1
 
 | | Crystal structure of protein BH3703 from Bacillus halodurans, Pfam DUF600 | | Descriptor: | BH3703 protein, GLYCEROL, SULFATE ION | | Authors: | Ramagopal, U.A, Russell, M, Toro, R, Freeman, J.C, Reyes, C, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of hypothetical protein BH3703 from Bacillus halodurans
To be Published
|
|
2HY3
 
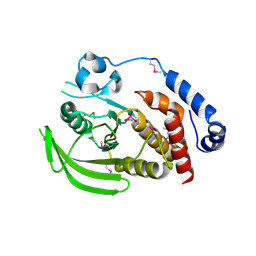 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2I5K
 
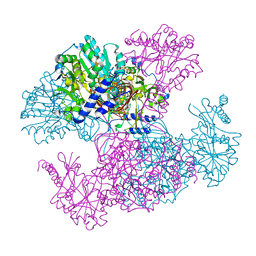 | | Crystal structure of Ugp1p | | Descriptor: | UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Roeben, A, Plitzko, J.M, Koerner, R, Boettcher, U.M.K, Siegers, K, Hayer-Hartl, M, Bracher, A. | | Deposit date: | 2006-08-25 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Subunit Assembly in UDP-glucose Pyrophosphorylase from Saccharomyces cerevisiae
J.Mol.Biol., 364, 2006
|
|
2I28
 
 | | Solution Structure of alpha-Conotoxin BuIA | | Descriptor: | Alpha-conotoxin BuIA | | Authors: | Chi, S.-W, Kim, D.-H, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of alpha-conotoxin BuIA, a novel neuronal nicotinic acetylcholine receptor antagonist with an unusual 4/4 disulfide scaffold
Biochem.Biophys.Res.Commun., 349, 2006
|
|
2IBK
 
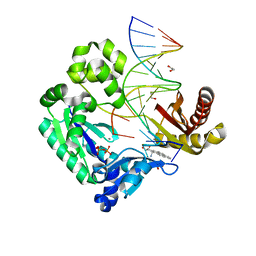 | | Bypass of Major Benzopyrene-dG Adduct by Y-Family DNA Polymerase with Unique Structural Gap | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*AP*T)-3', ... | | Authors: | Bauer, J, Ling, H, Sayer, J.M, Xing, G, Yagi, H, Jerina, D.M. | | Deposit date: | 2006-09-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I9I
 
 | | Crystal Structure of Helicobacter pylori protein HP0492 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Mckenzie, C, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-05 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Helicobacter pylori hypothetical protein O25234
TO BE PUBLISHED
|
|
2I5G
 
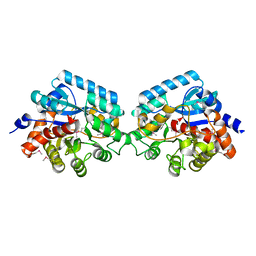 | | Crystal strcuture of amidohydrolase from Pseudomonas aeruginosa | | Descriptor: | amidohydrolase | | Authors: | Min, T, Sauder, J.M, Wasserman, S.R, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of amidohydrolase from Pseudomonas aeruginosa
To be Published
|
|
2I0O
 
 | | Crystal structure of Anopheles gambiae Ser/Thr phosphatase complexed with Zn2+ | | Descriptor: | Ser/Thr phosphatase, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2I52
 
 | | Crystal structure of protein PTO0218 from Picrophilus torridus, Pfam DUF372 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ramagopal, U.A, Gilmore, J, Toro, R, Bain, K.T, McKenzie, C, Reyes, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-23 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of hypothetical protein PTO0218 from Picrophilus torridus
To be Published
|
|
2D3O
 
 | | Structure of Ribosome Binding Domain of the Trigger Factor on the 50S ribosomal subunit from D. radiodurans | | Descriptor: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, ... | | Authors: | Schluenzen, F, Wilson, D.N, Hansen, H.A, Tian, P, Harms, J.M, McInnes, S.J, Albrecht, R, Buerger, J, Wilbanks, S.M, Fucini, P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Binding Mode of the Trigger Factor on the Ribosome: Implications for Protein Folding and SRP Interaction
Structure, 13, 2005
|
|
2IA6
 
 | | Bypass of Major Benzopyrene-dG Adduct by Y-Family DNA Polymerase with Unique Structural Gap | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*A)-3', ... | | Authors: | Bauer, J, Ling, H, Sayer, J.M, Xing, G, Yagi, H, Jerina, D.M. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2IHN
 
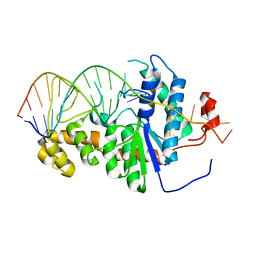 | | Co-crystal of Bacteriophage T4 RNase H with a fork DNA substrate | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*CP*C)-3', 5'-D(*GP*GP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*TP*AP*GP*TP*CP*AP*A)-3', Ribonuclease H | | Authors: | Devos, J.M, Mueser, T.C. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates.
J.Biol.Chem., 282, 2007
|
|
1YMT
 
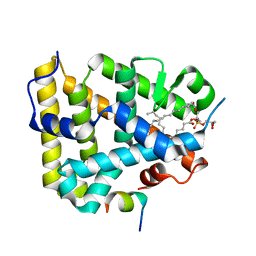 | | Mouse SF-1 LBD | | Descriptor: | 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL, Nuclear receptor 0B2, Steroidogenic factor 1 | | Authors: | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, Juzumiene, D, Bynum, J.M, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
8DKB
 
 | |
