6BXK
 
 | | Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and MTA | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Torelli, A.T, Fenwick, M.K, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXO
 
 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAH | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
8RZX
 
 | |
8T4P
 
 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and di-glutamate AGF347 inhibitor | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-{4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoyl}-L-gamma-glutamyl-L-glutamic acid, Serine hydroxymethyltransferase, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2023-06-09 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Characterization of 5-Substituted Pyrrolo[3,2- d ]pyrimidine Antifolate Inhibitors in Complex with Human Serine Hydroxymethyl Transferase 2.
Biochemistry, 2024
|
|
8T4O
 
 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and AGF347 inhibitor with no glutamate | | Descriptor: | 4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoic acid, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2023-06-09 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural Characterization of 5-Substituted Pyrrolo[3,2- d ]pyrimidine Antifolate Inhibitors in Complex with Human Serine Hydroxymethyl Transferase 2.
Biochemistry, 2024
|
|
8TLC
 
 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and tri-glutamate AGF347 inhibitor | | Descriptor: | N-{4-[4-(2-amino-4-oxo-1,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoyl}-D-gamma-glutamyl-L-gamma-glutamyl-D-glutamic acid, Serine hydroxymethyltransferase, mitochondrial | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Characterization of 5-Substituted Pyrrolo[3,2- d ]pyrimidine Antifolate Inhibitors in Complex with Human Serine Hydroxymethyl Transferase 2.
Biochemistry, 2024
|
|
4JLH
 
 | |
5Y97
 
 | | Crystal structure of snake gourd seed lectin in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Seed lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Chandran, T, Vijayan, M, Sivaji, N, Surolia, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ligand binding and retention in snake gourd seed lectin (SGSL). A crystallographic, thermodynamic and molecular dynamics study
Glycobiology, 28, 2018
|
|
4Z1R
 
 | | Crystal structure of collagen-like peptide at 1.27 Angstrom resolution | | Descriptor: | Collagen-like peptide | | Authors: | Plonska-Brzezinska, M.E, Czyrko, J, Brus, D.M, Imierska, M, Brzezinski, K. | | Deposit date: | 2015-03-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Triple helical collagen-like peptide interactions with selected polyphenolic compounds.
Rsc Adv, 5, 2015
|
|
5DM4
 
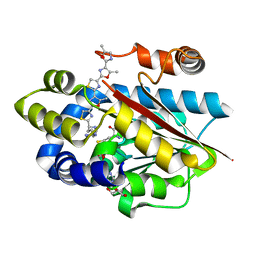 | | Crystal structure of the plantazolicin methyltransferase BpumL in complex with pentazolic desmethylPZN analog and SAH | | Descriptor: | 1-[(4S)-4-(4-{4-[4-(5,5'-dimethyl-2,4'-bi-1,3-oxazol-2'-yl)-1,3-thiazol-2-yl]-5-methyl-1,3-oxazol-2-yl}-1,3-thiazol-2-yl)-4-(methylamino)butyl]guanidine, GLYCEROL, Methyltransferase domain family, ... | | Authors: | Hao, Y, Nair, S.K. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into methyltransferase specificity and bioactivity of derivatives of the antibiotic plantazolicin.
Acs Chem.Biol., 10, 2015
|
|
2M9U
 
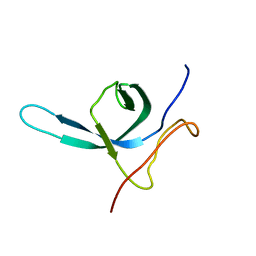 | | Solution NMR structure of the C-terminal domain (CTD) of Moloney murine leukemia virus integrase, Northeast Structural Genomics Target OR41A | | Descriptor: | Integrase p46 | | Authors: | Aiyer, S, Rossi, P, Schneider, W.M, Chander, A, Roth, M.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction.
Nucleic Acids Res., 42, 2014
|
|
5Y42
 
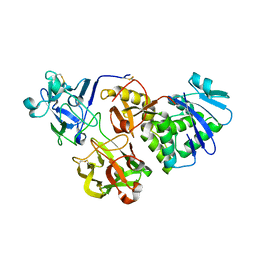 | |
5DLY
 
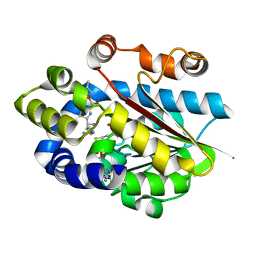 | |
5DM1
 
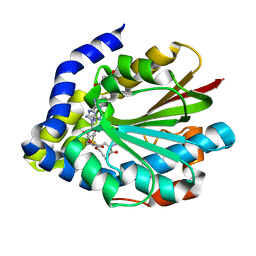 | |
5DM0
 
 | | Crystal structure of the plantazolicin methyltransferase BamL in complex with triazolic desmethylPZN analog and SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ethyl 2-(2-{2-[(1S)-1-amino-4-carbamimidamidobutyl]-1,3-thiazol-4-yl}-5-methyl-1,3-oxazol-4-yl)-1,3-thiazole-4-carboxylate, ... | | Authors: | Hao, Y, Nair, S.K. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into methyltransferase specificity and bioactivity of derivatives of the antibiotic plantazolicin.
Acs Chem.Biol., 10, 2015
|
|
5DM2
 
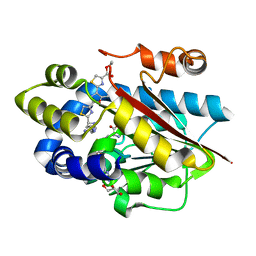 | |
3Q9A
 
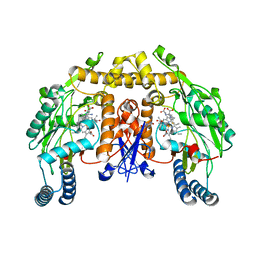 | | Structure of neuronal nitric oxide synthase in the ferric state in complex with N-5-[2-(ethylsulfanyl)ethanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Doukov, T, Poulos, T.L. | | Deposit date: | 2011-01-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Temperature-dependent spin crossover in neuronal nitric oxide synthase bound with the heme-coordinating thioether inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
3Q99
 
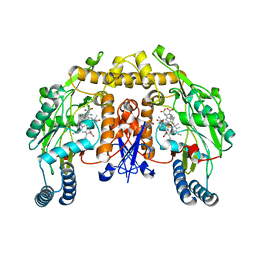 | | Structure of neuronal nitric oxide synthase in the ferric state in complex with N~5~-[(3-(ethylsulfanyl)propanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Doukov, T, Poulos, T.L. | | Deposit date: | 2011-01-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Temperature-dependent spin crossover in neuronal nitric oxide synthase bound with the heme-coordinating thioether inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
2BD1
 
 | | A possible role of the second calcium ion in interfacial binding: Atomic and medium resolution crystal structures of the quadruple mutant of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Phospholipase A2 | | Authors: | Sekar, K, Velmurugan, D, Tsai, M.D. | | Deposit date: | 2005-10-19 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Suggestive evidence for the involvement of the second calcium and surface loop in interfacial binding: monoclinic and trigonal crystal structures of a quadruple mutant of phospholipase A(2).
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BCH
 
 | | A possible of Second calcium ion in interfacial binding: Atomic and Medium resolution crystal structures of the quadruple mutant of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Poi, M.J, Dauter, Z, Tsai, M.D. | | Deposit date: | 2005-10-19 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Suggestive evidence for the involvement of the second calcium and surface loop in interfacial binding: monoclinic and trigonal crystal structures of a quadruple mutant of phospholipase A(2).
Acta Crystallogr.,Sect.D, 62, 2006
|
|
