2QHK
 
 | |
2QM3
 
 | | Crystal structure of a predicted methyltransferase from Pyrococcus furiosus | | Descriptor: | ACETIC ACID, CALCIUM ION, Predicted methyltransferase | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a predicted methyltransferase from Pyrococcus furiosus.
TO BE PUBLISHED
|
|
2QSW
 
 | | Crystal structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis | | Descriptor: | GLYCEROL, Methionine import ATP-binding protein metN 2, ZINC ION | | Authors: | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis.
To be Published
|
|
3M6Y
 
 | | Structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at 1.45 a resolution. | | Descriptor: | 4-Hydroxy-2-oxoglutarate aldolase, CALCIUM ION, CHLORIDE ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of 4-Hydroxy-2-Oxoglutarate Aldolase from Bacillus Cereus at 1.45 A Resolution.
To be Published
|
|
3O5V
 
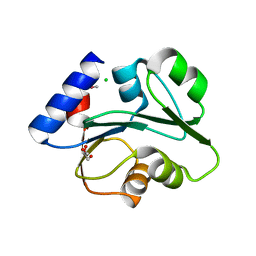 | | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A | | Descriptor: | CHLORIDE ION, GLYCEROL, X-PRO dipeptidase | | Authors: | Stein, A.J, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A
To be Published
|
|
6S44
 
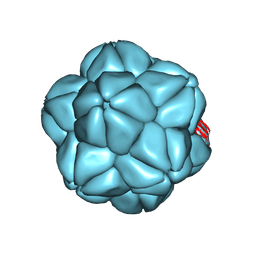 | |
3OBF
 
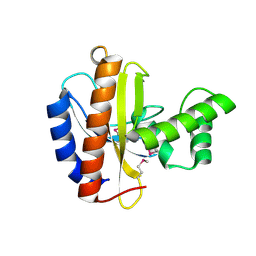 | | Crystal structure of putative transcriptional regulator, IclR family; targeted domain 129...302 | | Descriptor: | Putative transcriptional regulator, IclR family | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of putative transcriptional regulator, IclR family; targeted domain 129...302
To be Published
|
|
3OJ0
 
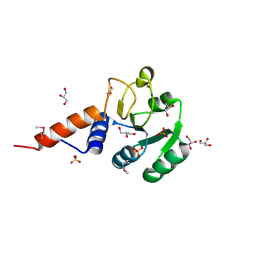 | | Crystal structure of glutamyl-tRNA reductase from Thermoplasma volcanium (nucleotide binding domain) | | Descriptor: | GLYCEROL, Glutamyl-tRNA reductase, SULFATE ION | | Authors: | Michalska, K, Marshall, N, Clancy, S, Puttagunta, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Crystal structure of glutamyl-tRNA reductase from Thermoplasma volcanium (nucleotide binding domain)
To be Published
|
|
3OOO
 
 | | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V | | Descriptor: | Proline dipeptidase | | Authors: | Fan, Y, Wu, R, Morales, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V
To be Published
|
|
3OVK
 
 | |
5N8N
 
 | | Contracted sheath of a Pseudomonas aeruginosa type six secretion system consisting of TssB1 and TssC1 | | Descriptor: | EvpB family type VI secretion protein, Type VI secretion protein, family | | Authors: | Salih, O, He, S, Stach, L, Macdonald, J.T, Planamente, S, Manoli, E, Scheres, S, Filloux, A, Freemont, P.S. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Atomic Structure of Type VI Contractile Sheath from Pseudomonas aeruginosa.
Structure, 26, 2018
|
|
3MZY
 
 | |
3P4F
 
 | | Structural and biochemical insights into MLL1 core complex assembly and regulation. | | Descriptor: | Histone-lysine N-methyltransferase MLL, Retinoblastoma-binding protein 5, WD repeat-containing protein 5 | | Authors: | Avdic, V, Zhang, P, Lanouette, S, Groulx, A, Tremblay, V, Brunzelle, J.B, Couture, J.-F. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical insights into MLL1 core complex assembly.
Structure, 19, 2011
|
|
4RGA
 
 | | Phage 1358 receptor binding protein in complex with the trisaccharide GlcNAc-Galf-GlcOMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-galactofuranose-(1-6)-methyl alpha-D-glucopyranoside, Phage 1358 receptor binding protein (ORF20) | | Authors: | Spinelli, S, Mccabe, O, Farenc, C, Tremblay, D, Blangy, S, Oscarson, S, Moineau, S, Cambillau, C. | | Deposit date: | 2014-09-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors.
Mol.Microbiol., 96, 2015
|
|
3MVN
 
 | | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP | | Descriptor: | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP
To be Published
|
|
4O2H
 
 | | Crystal structure of BCAM1869 protein (RsaM homolog) from Burkholderia cenocepacia | | Descriptor: | protein BCAM1869 | | Authors: | Michalska, K, Chhor, G, Clancy, S, Winans, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-22 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RsaM: a transcriptional regulator of Burkholderia spp. with novel fold.
Febs J., 281, 2014
|
|
3MKL
 
 | |
3MR7
 
 | |
3OOS
 
 | | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne | | Descriptor: | Alpha/beta hydrolase family protein, GLYCEROL, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Bigelow, L, Hamilton, J, Li, H, Zhou, Y, Clancy, S, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne
To be Published
|
|
3PEB
 
 | | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Dipeptidase, ... | | Authors: | Cuff, M.E, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus.
TO BE PUBLISHED
|
|
3PFM
 
 | | Crystal structure of a EAL domain of GGDEF domain protein from Pseudomonas fluorescens Pf | | Descriptor: | GGDEF domain protein | | Authors: | Nocek, B, Stein, A, Marshall, N, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Crystal structure of a EAL domain of GGDEF domain protein from Pseudomonas fluorescens Pf
TO BE PUBLISHED
|
|
3N2Q
 
 | |
3NE7
 
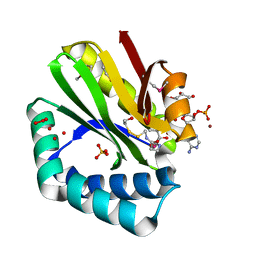 | | Crystal structure of paia n-acetyltransferase from thermoplasma acidophilum in complex with coenzyme a | | Descriptor: | ACETYLTRANSFERASE, BETA-MERCAPTOETHANOL, COENZYME A, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
3C8G
 
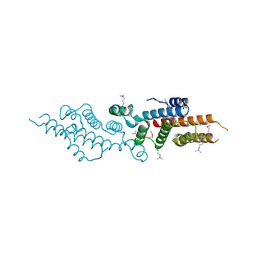 | | Crystal structure of a possible transciptional regulator YggD from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, Putative transcriptional regulator | | Authors: | Tan, K, Borovilos, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
