1S45
 
 | | Crystal structure analysis of the DNA quadruplex d(TGGGGT) S1 | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Caceres, C, Wright, G, Gouyette, C, Parkinson, G, Subirana, J.A. | | Deposit date: | 2004-01-15 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+1/Na+1 ions
Nucleic Acids Res., 32, 2004
|
|
5D0J
 
 | | Grb7 SH2 with inhibitor peptide | | Descriptor: | G7-TEdFP peptide, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Gunzburg, M.J, Watson, G.M, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2015-08-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
3ZBU
 
 | | Ferredoxin-NADP Reductase Mutant with SER 80 Replaced by ALA (S80A) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
1S5W
 
 | |
1S69
 
 | | The X-ray structure of the cyanobacteria Synechocystis hemoglobin "cyanoglobin" with cyanide ligand | | Descriptor: | CITRATE ANION, CYANIDE ION, Cyanoglobin, ... | | Authors: | Trent III, J.T, Kundu, S, Hoy, J.A, Hargrove, M.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic analysis of synechocystis cyanoglobin reveals the structural changes accompanying ligand binding in a hexacoordinate hemoglobin.
J.Mol.Biol., 341, 2004
|
|
1SCE
 
 | |
3CTW
 
 | |
3ZVU
 
 | | Structure of the PYR1 His60Pro mutant in complex with the HAB1 phosphatase and Abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABSCISIC ACID RECEPTOR PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thermodynamic Switch Modulates Abscisic Acid Receptor Sensitivity.
Embo J., 30, 2011
|
|
3D4M
 
 | | Glutaredoxin 2 oxidized structure | | Descriptor: | Glutaredoxin-2, mitochondrial | | Authors: | Discola, K.F, de Oliveira, M.A, Barcena, J.A, Porras, P, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2008-05-14 | | Release date: | 2008-10-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae.
J.Mol.Biol., 385, 2009
|
|
3CKE
 
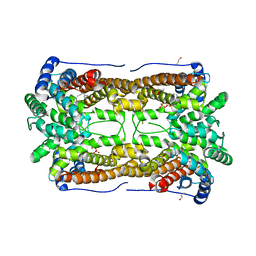 | | Crystal structure of aristolochene synthase in complex with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Yu, F, Miller, D.J, Faraldos, J.A, Zhao, Y, Coates, R.M, Allemann, R.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis.
J.Biol.Chem., 283, 2008
|
|
3CMH
 
 | | SYNTHETIC LINEAR TRUNCATED ENDOTHELIN-1 AGONIST | | Descriptor: | PROTEIN (ENDOTHELIN-1) | | Authors: | Hewage, C.M, Jiang, L, Parkinson, J.A, Ramage, R, Sadler, I.H. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel ETB receptor selective agonist ET1-21 [Cys(Acm)1,15, Aib3,11, Leu7] by nuclear magnetic resonance spectroscopy and molecular modelling.
J.Pept.Res., 53, 1999
|
|
5AMW
 
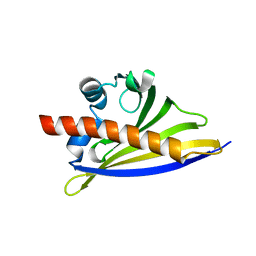 | | Crystal Structure of the Strawberry Pathogenesis-Related 10 (PR-10) Fra a 2 protein (A141F) processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | Fra a 2 allergen | | Authors: | Zander, U, Casanal, A, Pott, D, Valpuesta, V, Hoffmann, G, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
7JRO
 
 | | Plant Mitochondrial complex IV from Vigna radiata | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CARDIOLIPIN, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of respiratory complex III 2 , complex IV, and supercomplex III 2 -IV from vascular plants.
Elife, 10, 2021
|
|
5ANK
 
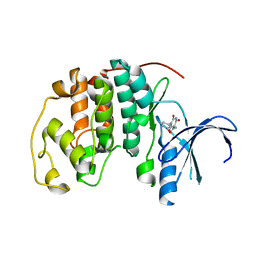 | | Crystal structure of CDK2 in complex with 2,4,6-trioxo-1-phenyl- hexahydropyrimidine-5-carboxamide processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | 2,4,6-TRIOXO-1-PHENYL-HEXAHYDROPYRIMIDINE-5-CARBOXAMIDE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5AYI
 
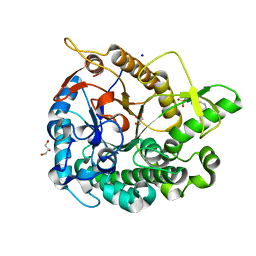 | | Crystal structure of GH1 Beta-glucosidase TD2F2 N223Q mutant | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-GLUCOSIDASE, GLYCEROL, ... | | Authors: | Jo, T, Manninen, J.A, Matsuzawa, T, Uchiyama, T, Yaoi, K, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-08-21 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and identification of a key amino acid for glucose tolerance, substrate specificity, and transglycosylation activity of metagenomic beta-glucosidase Td2F2
Febs J., 283, 2016
|
|
3CQP
 
 | | Human SOD1 G85R Variant, Structure I | | Descriptor: | COPPER (II) ION, MALONATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cao, X, Antonyuk, S, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
3V2T
 
 | |
3CTL
 
 | | Crystal structure of D-Allulose 6-Phosphate 3-Epimerase from Escherichia coli K12 complexed with D-glucitol 6-phosphate and magnesium | | Descriptor: | D-SORBITOL-6-PHOSPHATE, D-allulose-6-phosphate 3-epimerase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-04-14 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate specificity in phosphate binding (beta/alpha)8-barrels: D-allulose 6-phosphate 3-epimerase from Escherichia coli K-12.
Biochemistry, 47, 2008
|
|
5AMY
 
 | |
1RMK
 
 | | Solution structure of conotoxin MrVIB | | Descriptor: | Mu-O-conotoxin MrVIB | | Authors: | Daly, N.L, Ekberg, J.A, Thomas, L, Adams, D.J, Lewis, R.J, Craik, D.J. | | Deposit date: | 2003-11-28 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structures of muO-conotoxins from Conus marmoreus. Inhibitors of tetrodotoxin (TTX)-sensitive and TTX-resistant sodium channels in mammalian sensory neurons
J.Biol.Chem., 279, 2004
|
|
1RN9
 
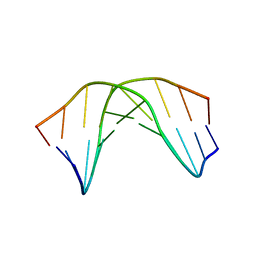 | | A SHORT LEXITROPSIN THAT RECOGNIZES THE DNA MINOR GROOVE AT 5'-ACTAGT-3': UNDERSTANDING THE ROLE OF ISOPROPYL-THIAZOLE | | Descriptor: | 5'-D(*CP*GP*AP*CP*TP*AP*GP*TP*CP*G)-3' | | Authors: | Anthony, N.G, Johnston, B.F, Khalaf, A.I, Mackay, S.P, Parkinson, J.A, Suckling, C.J, Waigh, R.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-08-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Short Lexitropsin that Recognizes the DNA Minor Groove at 5'-ACTAGT-3': Understanding the Role of Isopropyl-thiazole.
J.Am.Chem.Soc., 126, 2004
|
|
3D0O
 
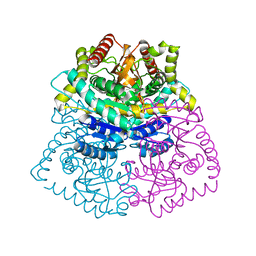 | |
1T9B
 
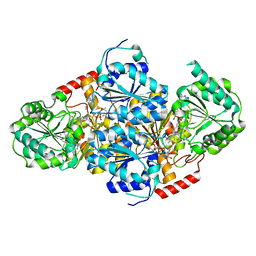 | | Crystal structure of yeast acetohydroxyacid synthase in complex with a sulfonylurea herbicide, chlorsulfuron | | Descriptor: | 1-(2-CHLOROPHENYLSULFONYL)-3-(4-METHOXY-6-METHYL-L,3,5-TRIAZIN-2-YL)UREA, 2-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}PROPANE-1-THIOL, 5-(AMINOMETHYL)-2-METHYLPYRIMIDIN-4-AMINE, ... | | Authors: | McCourt, J.A, Pang, S.S, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2004-05-16 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidating the specificity of binding of sulfonylurea herbicides to acetohydroxyacid synthase.
Biochemistry, 44, 2005
|
|
1TA2
 
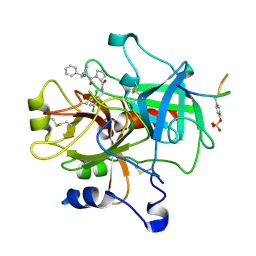 | | Crystal structure of thrombin in complex with compound 1 | | Descriptor: | 1-(2-AMINO-3,3-DIPHENYL-PROPIONYL)-PYRROLIDINE-3-CARBOXYLIC ACID 2,5-DICHLORO-BENZYLAMIDE, Hirudin, thrombin | | Authors: | Tucker, T.J, Brady, S.F, Lumma, W.C, Lewis, S.D, Gardel, S.J, Naylor-Olsen, A.M, Yan, Y, Sisko, J.T, Stauffer, K.J, Lucas, B.Y, Lynch, J.J, Cook, J.J, Stranieri, M.T, Holahan, M.A, Lyle, E.A, Baskin, E.P, Chen, I.-W, Dancheck, K.B, Krueger, J.A, Cooper, C.M, Vacca, J.P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-06-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of a series of potent and orally bioavailable
noncovalent thrombin inhibitors that utilize nonbasic groups in the P1 position
J.Med.Chem., 41, 1998
|
|
3ZXN
 
 | | Moorella thermoacetica RsbS S58E | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN, THIOCYANATE ION | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-08-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
