8I4W
 
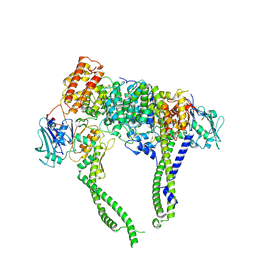 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2023-01-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6LR0
 
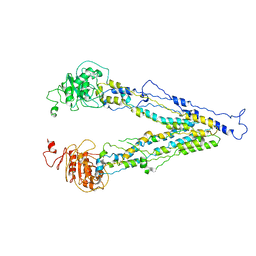 | | structure of human bile salt exporter ABCB11 | | Descriptor: | Bile salt export pump | | Authors: | Wang, L, Hou, W.T, Chen, L, Jiang, Y.L, Xu, D, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human bile salts exporter ABCB11.
Cell Res., 30, 2020
|
|
5KDT
 
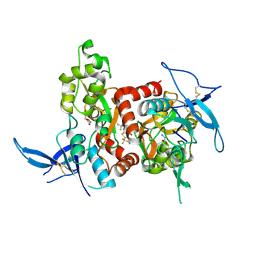 | | Structure of the human GluN1/GluN2A LBD in complex with GNE0723 | | Descriptor: | (1~{R},2~{R})-2-[7-[[5-chloranyl-3-(trifluoromethyl)pyrazol-1-yl]methyl]-5-oxidanylidene-2-(trifluoromethyl)-[1,3]thiazolo[3,2-a]pyrimidin-3-yl]cyclopropane-1-carbonitrile, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J. Med. Chem., 59, 2016
|
|
1AGW
 
 | |
8IBY
 
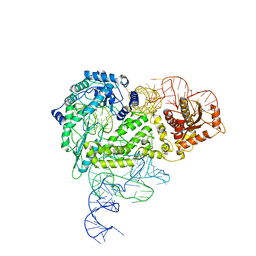 | | Structure of R2 with 5'ORF | | Descriptor: | 5'ORF RNA, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
6LKX
 
 | | The structure of PRRSV helicase | | Descriptor: | CITRIC ACID, GLYCEROL, RNA-dependent RNA polymerase, ... | | Authors: | Shi, Y.J, Tong, X.H, Peng, G.Q. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural Characterization of the Helicase nsp10 Encoded by Porcine Reproductive and Respiratory Syndrome Virus.
J.Virol., 94, 2020
|
|
3PWY
 
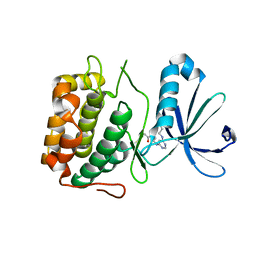 | | Crystal structure of an extender (SPD28345)-modified human PDK1 complex 2 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, N-[2-({6-[(2-sulfanylethyl)amino]pyrimidin-4-yl}amino)ethyl]propanamide | | Authors: | Elling, R.A, Penny, D.M, Simmons, R.L, Erlanson, D.A, Romanowski, M.J. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QC4
 
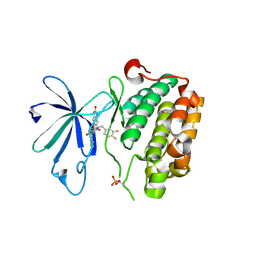 | | PDK1 in complex with DFG-OUT inhibitor xxx | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Arndt, J.W. | | Deposit date: | 2011-01-15 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6O76
 
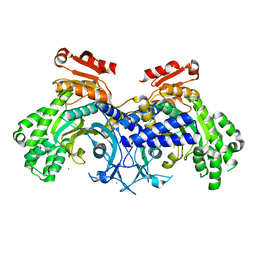 | | Human cytosolic Histidyl-tRNA synthetase (HisRS) with WHEP domain | | Descriptor: | CHLORIDE ION, Histidine--tRNA ligase, cytoplasmic | | Authors: | Kuhle, B, Yang, X.L. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | CMT disease severity correlates with mutation-induced open conformation of histidyl-tRNA synthetase, not aminoacylation loss, in patient cells.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7URS
 
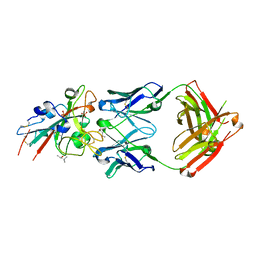 | |
7URQ
 
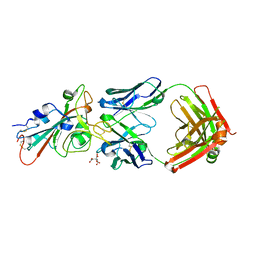 | |
1FGI
 
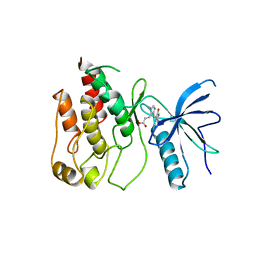 | |
8TQP
 
 | | HIV-CA Disulfide linked Hexamer bound to Quinazolin-4-one Scaffold inhibitor | | Descriptor: | 2-[4-(4-aminobenzene-1-sulfonyl)-2-oxopiperazin-1-yl]-N-{(1R)-2-(3,5-difluorophenyl)-1-[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]ethyl}acetamide, Gag polyprotein | | Authors: | Goldstone, D.C, Barnett, M.J, Taka, J.R.H. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery, Crystallographic Studies, and Mechanistic Investigations of Novel Phenylalanine Derivatives Bearing a Quinazolin-4-one Scaffold as Potent HIV Capsid Modulators.
J.Med.Chem., 66, 2023
|
|
8TOV
 
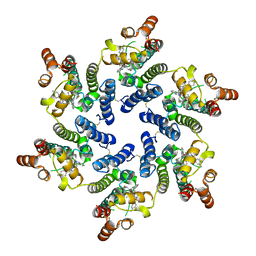 | | HIV-CA Disulfide linked Hexamer bound to Quinazolin-4-one Scaffold inhibitor | | Descriptor: | 2-[4-(4-aminobenzene-1-sulfonyl)-2-oxopiperazin-1-yl]-N-[(1R)-2-(3,5-difluorophenyl)-1-{3-[4-(morpholine-4-sulfonyl)phenyl]-4-oxo-3,4-dihydroquinazolin-2-yl}ethyl]acetamide, Matrix protein p17 | | Authors: | Goldstone, D.C, Barnett, M.J, Taka, J.R.H. | | Deposit date: | 2023-08-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, Crystallographic Studies, and Mechanistic Investigations of Novel Phenylalanine Derivatives Bearing a Quinazolin-4-one Scaffold as Potent HIV Capsid Modulators.
J.Med.Chem., 66, 2023
|
|
8T9R
 
 | |
8T1X
 
 | |
9CL0
 
 | |
9CKZ
 
 | | Cryo-EM structure of human XPR1 in apo state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Wang, Z, Wu, H. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | The identification of XPR1 as a voltage- and phosphate-activated phosphate-permeable ion channel.
Nat Commun, 16, 2025
|
|
5I2K
 
 | | Structure of the human GluN1/GluN2A LBD in complex with 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 19) | | Descriptor: | 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
5I9E
 
 | | Crystal structure of a nuclear actin ternary complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Chen, Z, Cao, T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a nuclear actin ternary complex.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9B83
 
 | |
9B84
 
 | |
9B89
 
 | |
